胰腺癌(pancreatic cancer)一种预后极差的消化道恶性肿瘤,常指胰腺导管腺癌,占胰腺恶性肿瘤的95%以上[1]。近年来胰腺癌外科技术日臻完善,围手术期病死率和术后并发症发生率均有显著下降,但是,其总体手术切除率和远期生存率的提高并不明显。究其关键原因所在,胰腺癌的解剖部位特殊及其复杂的肿瘤生物学行为,导致进展迅速,预后不佳[2]。研究表明,胰腺癌由最初的微观的非侵入性上皮增生胰腺管,进展到晚期,均离不开原癌基因的驱动[3],诸如表皮生长因子受体(EGFR)及乳酸脱氢酶(LDHA)参与调控胰腺癌细胞代谢[4],介导肿瘤局部免疫微环境异常[5],促进辅助化疗与吉西他滨类药物治疗抵抗[6-7]。目前对于EGFR和LDHA在胰腺癌局部免疫微环境中的作用及相关的分子机制仍知之甚少。随着肿瘤免疫基因组学(immunogenomics)的进展,可以试图通过TIMER(Tumor IMmune Estimation Resource)数据库的研究[8],找到对胰腺癌起到关键作用的驱动因子与之相对应的重要信号通道。因此笔者通过TIMER数据库结合人类蛋白表达图谱分析了EGFR及LDHA mRNA和蛋白在胰腺癌中的表达信息,以期为进一步研究胰腺癌发生发展中的作用机制提供有价值的佐证。
1 资料与方法
1.1 一般资料
TIMER数据库原发性胰腺癌组织RNA片段(RNA-seq)测序表达数据,共计178例。白种人157例,黑种人6例,其他15例;男98例,女80例;年龄(64.64±10.93)岁;肿瘤直径0.3~14 cm,平均(3.873±1.078)cm;局部侵润177例,无侵润1例;肿瘤分化程度G1级31例、G2级95例、G3级48例、G4级2例、其他2例;区域淋巴结转移N049例、N1120例、N1b 4例、Nd 1例、Nx 4例;无远处转移(M0)80例、有远处转移(M1)4例、无法评价有无远处转移(Mx)94例。TNM分期按AJCC第6版标准:IA 5例、IB 15例、IIA 28例、IIB 119例、III 4例、IV 4例、其他未分期3例。
1.2 TIMER数据库提取数据
运行TIMER数据库(https://cistrome.shinyapps.io/timer)搜索EGFR和LDHA在178例胰腺癌组织中与正常的胰腺组织的差异表达情况、临床预后、体细胞拷贝数变异(somatic CNA)、基因表达相关性。Cox比例风险模型协变量:年龄、性别、种族、第6版ACJJ肿瘤TNM分期(II、III、IV期)、肿瘤纯度(tumor purity)、免疫细胞浸润(CD4+T细胞、巨噬细胞、树状突细胞)和基因表达。患者总体生存率预测设置范围为5%~50%。通过拟合函数包R语言包绘制生存曲线。
1.3 人类蛋白表达图谱组织芯片数据库提取数据
运用人类蛋白表达图谱数据库(http://www.proteinatlas.org)搜索EGFR、LDHA、P38、FOXM1、CSF1和CSF1R蛋白在胰腺癌组织亚细胞定位(细胞浆/膜/核);并与正常的胰腺组织中的表达丰度相比较。
1.4 统计学处理
EGFR和LDHA mRNA表达水平用均数±标准差( ±s),两者表达相关性采用Pearson相关系数相分析。TIMER数据库相关性分析方法应用Spearman等级相关性分析。患者生存率采用Kaplan-Meier法计算,Log-rank法比较总体生存率的差异。Cox回归模型H R设定95%可信区间(CI)。界定P<0.05为差异有统计学意义。
±s),两者表达相关性采用Pearson相关系数相分析。TIMER数据库相关性分析方法应用Spearman等级相关性分析。患者生存率采用Kaplan-Meier法计算,Log-rank法比较总体生存率的差异。Cox回归模型H R设定95%可信区间(CI)。界定P<0.05为差异有统计学意义。
2 结 果
2.1 EGFR和LDHA在胰腺癌中及其之间的关系
EGFRm RNA在胰腺癌中的表达水平为4.96±1.53,在正常胰腺组织中的表达水平2.15±1.34;LDHA mRNA在胰腺癌中的表达水平为12.35±2.06,正常胰腺组织中的表达水平4.53±2.32;两者在胰腺癌组织中的表达水平均明显高于正常胰腺组织(均P<0.05)(图1)。Pearson相关系数相关性分析结果显示(图2),EGFR和LDHA mRNA表达呈明显正相关(r=0.511,P<0.001)。
通过TIMER数据库进行Spear man等级相关性分析结果显示,在胰腺癌局部免疫微环境中,EGFR和LDHA mRNA表达呈明显正相关(P<0.001)(图3)。
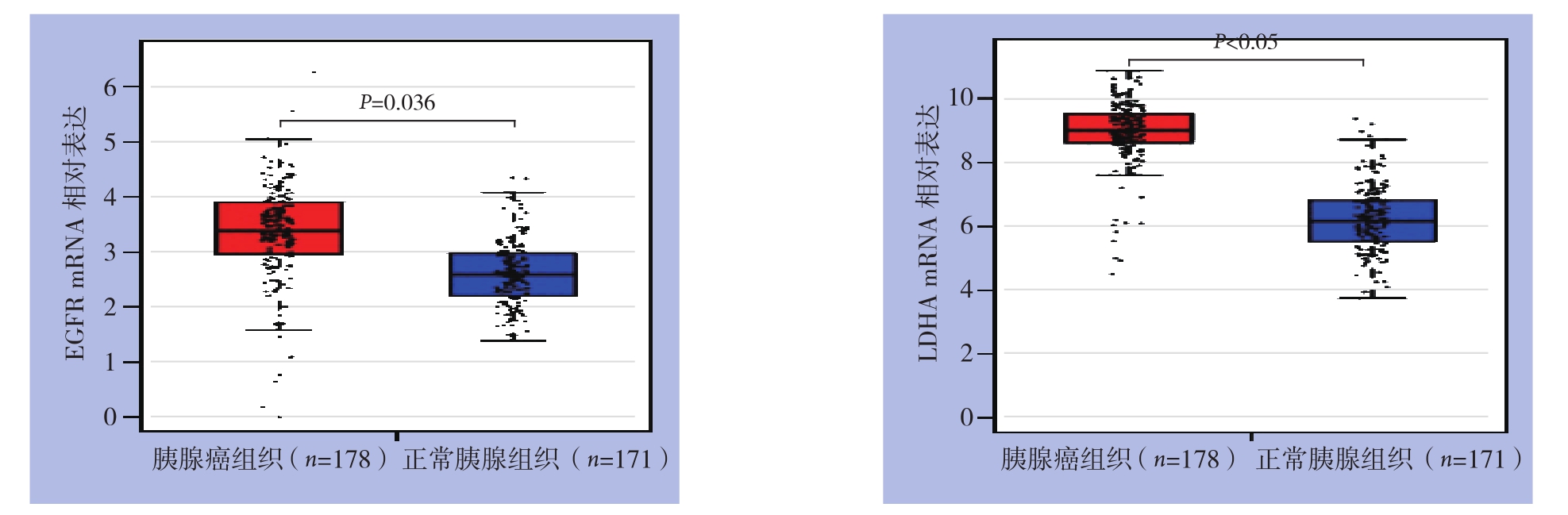
图1 EGFR与LDHA mRNA在胰腺癌和正常胰腺组织中的表达
Figure1 The mRNA expression levels of EGFR and LDHA in pancreatic cancer and normal pancreatic tissues
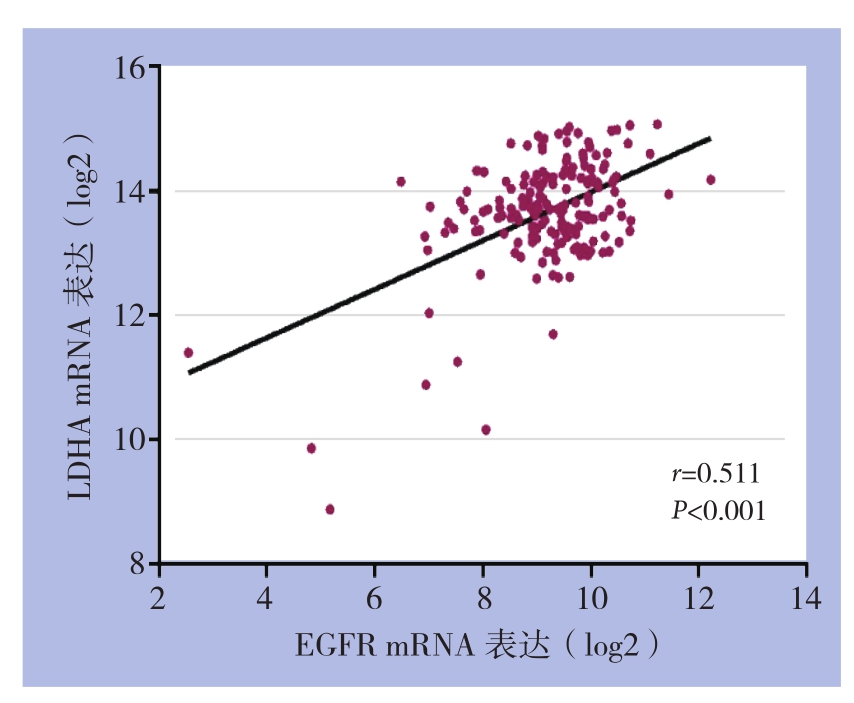
图2 胰腺癌组织中EGFR和LDHA mRNA表达的相关性分析
Figure2 Correlation analysis of between EGFR and LDHA mRNA expressions in pancreatic cancer
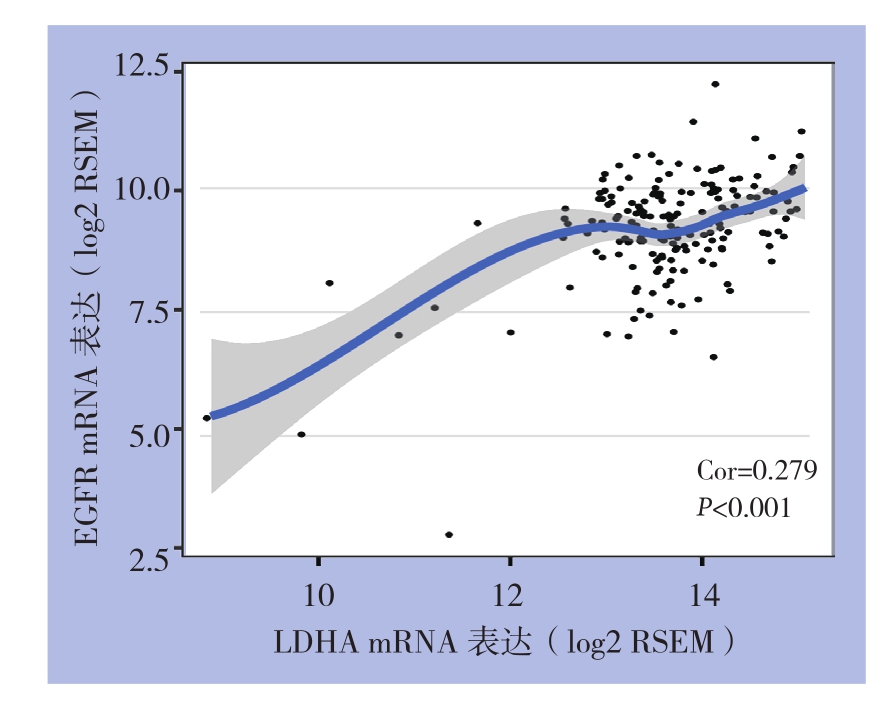
图3 TIMER数据库EGFR和LDHA mRNA表达的相关性分析
Figure3 Correlation analysis of between EGFR and LDHA mRNA expressions based on TIMER database
2.2 EGFR和LDHA mRNA高表达与胰腺癌预后的关系
TIMER数据库收集178例胰腺癌患者基因表达及总生存期数据,根据设定中位表达量35%,分为高表达及低表达两组,预测两组患者间的生存期差异,R语言绘制Kaplan-Meier生存曲线。结果提示巨噬细胞、树状突细胞高表达及低表达胰腺癌患者的1、3、5年生存率间无统计学差异(均P>0.05),而CD4+T细胞、EGFR和LDHA mRNA高表达及低表达胰腺癌患者的1、3、5年生存率间差异有统计学意义(P=0.042、0.013、0.001)。CD4+T细胞高表达预后好于其低表达;EGFR和LDHA低表达患者预后好于其高表达(图4)。
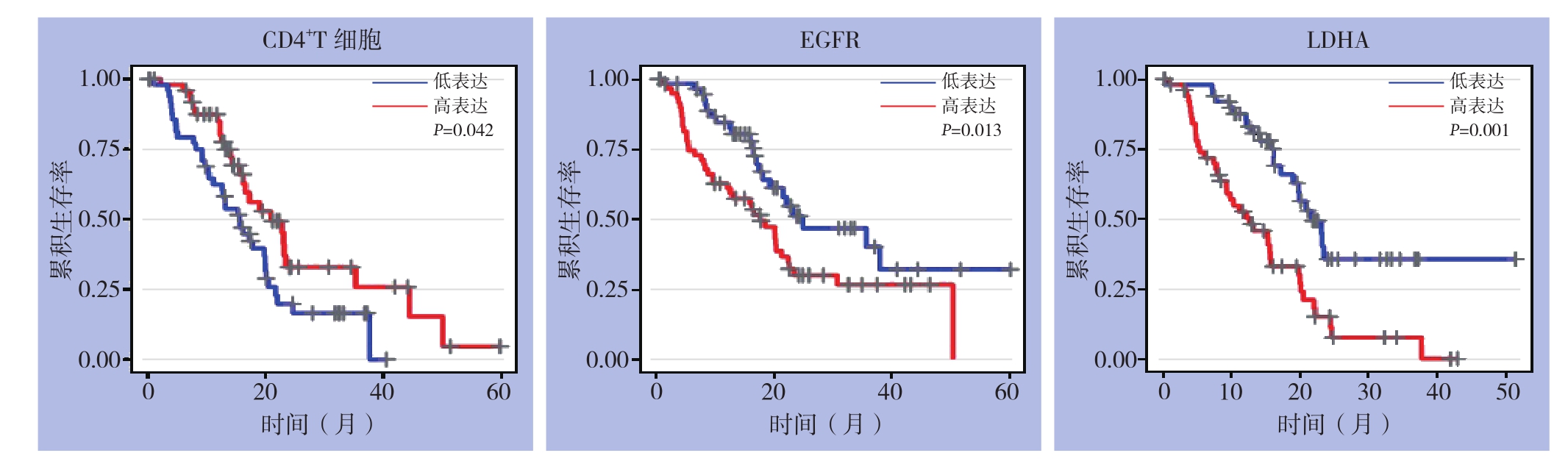
图4 TIMER数据库胰腺癌患者总体生存期分析
Figure4 Analysis of the overall survival of pancreatic cancer patients from TIMER database
2.3 胰腺癌预后影响因素的Cox比例风险回归模型结果
Cox比例风险回归模型分析结果显示:年龄(P=0.005)、肿瘤纯度(P=0.042)和LDHA(P<0.001)是影响胰腺癌预后的独立危险因素。而TNM分期、CD4+T细胞、巨噬细胞、树状突细胞浸润、EGFR表达水平非胰腺癌预后独立危险因素(均P>0.05)(表1)。
表1 胰腺癌患者总体生存率影响因素的Cox比例风险回归模型分析
Table1 Analysis of influential factors for overall survival of pancreatic cancer patients by using Cox proportional hazard model
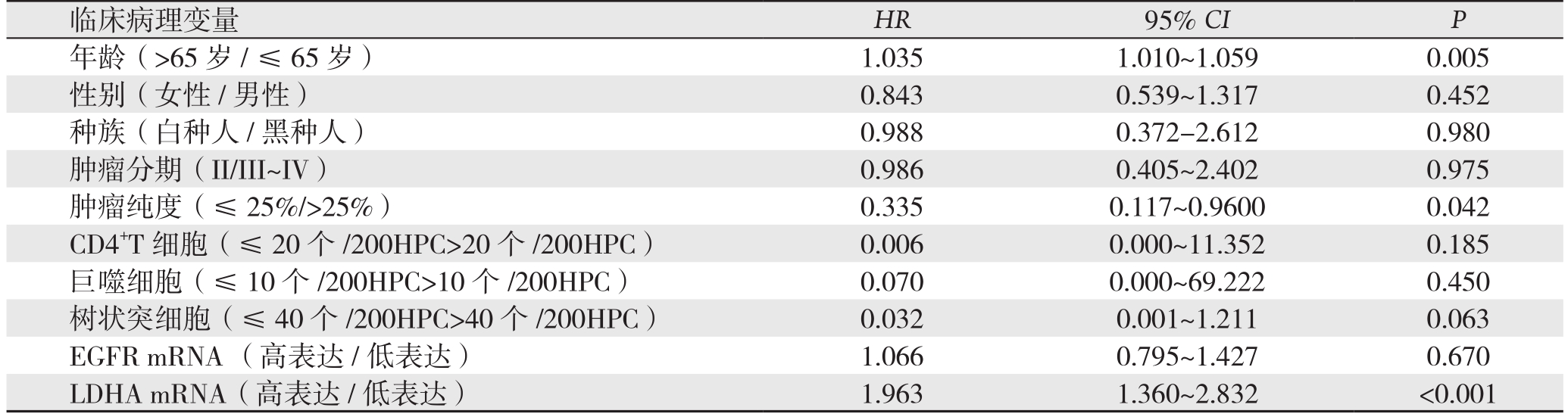
临床病理变量HR 95% CI P年龄(>65岁/≤65岁) 1.035 1.010~1.059 0.005性别(女性/男性) 0.843 0.539~1.317 0.452种族(白种人/黑种人) 0.988 0.372-2.612 0.980肿瘤分期(II/III~IV) 0.986 0.405~2.402 0.975肿瘤纯度(≤ 25%/>25%) 0.335 0.117~0.9600 0.042 CD4+T细胞(≤20个/200HPC>20个/200HPC) 0.006 0.000~11.352 0.185巨噬细胞(≤10个/200HPC>10个/200HPC) 0.070 0.000~69.222 0.450树状突细胞(≤40个/200HPC>40个/200HPC) 0.032 0.001~1.211 0.063 EGFR mRNA (高表达 /低表达) 1.066 0.795~1.427 0.670 LDHA mRNA(高表达/低表达) 1.963 1.360~2.832 <0.001
2.4 胰腺癌信号通路分子相关性表达分析
基于文献P38[9]、 FOXM1[10]、 PD-L1[11]、CSF1和CSF1R[12]报道在胰腺癌发生发展中的重要作用。通过TIMER数据库进行Spearman等级相关性分析(图5),结果显示:EGFR与P38、FOXM1、PD-L1、CSF1和CSF1R表达呈明显正相关(均P<0.001);而LDHA与FOXM1、CSF1和CSF1R表达呈明显正相关(均P<0.001)。
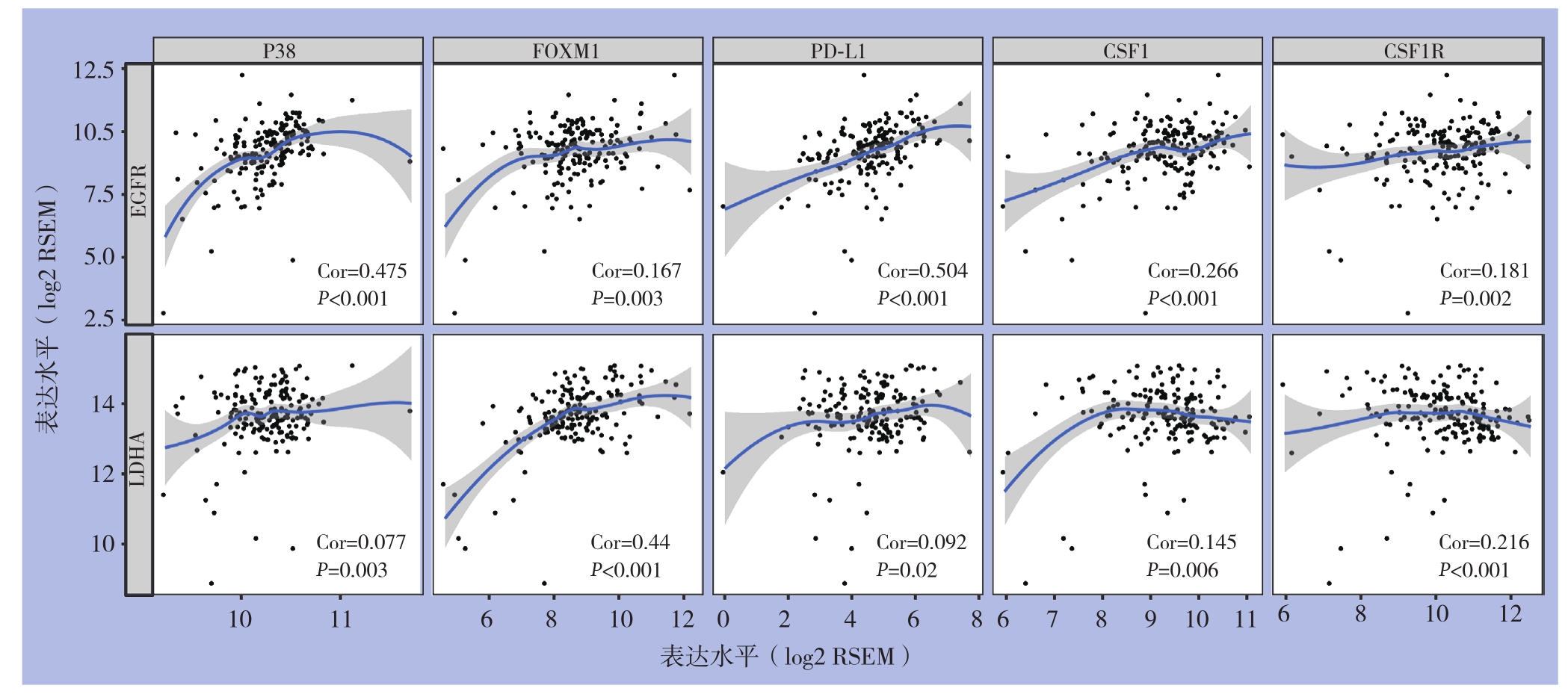
图5 EGFR、LDHA与胰腺癌相关分子mRNA表达的相关性分析
Figure5 Correlation analysis of EGFR and LDHA with the mRNA expressions in the molecules associated with pancreatic cancer
2.5 EGFR、LDHA及胰腺癌信号通路分子在胰腺癌组织中的表达
为验证EGFR、LDHA、P38、FOXM1和CSF1R mRNA在胰腺癌组织中的蛋白表达及定位。通过人类蛋白表达图谱数据库检索结果显示,EGFR、LDHA、P38、FOXM1和CSF1R蛋白在胰腺癌组织亚细胞定位细胞浆或者细胞膜;并且表达丰度高于正常胰腺组织(图6)。

图6 EGFR、LDHA、P38、FOXM1和CSF1R蛋白在正常胰腺与胰腺癌组织中的表达
Figure6 The protein expression patterns of EGFR, LDHA, P38, FOXM1 and CSF1R in pancreas cancer and normal pancreatic tissues
3 讨 论
研究表明,利用生物信息学技术,分析和处理胰腺癌相关高通量数据,整合基因组[13]、单核苷酸多态性(SNP)[14]、miRNA[15]、蛋白质调控网络以及通路功能注释等信息数据,可以迅速准确地分析某一个或多个基因在特定肿瘤中的表达差异;例如胰腺癌相关基因(TP53、SMAD4、CDKN2A、ARID1A、ROBO2)、胰腺癌候选驱动基因KDM6A和PREX2;并对胰腺癌的作用机制及调控分子网络进行研究[16],探讨胰腺癌肿瘤异质性和肿瘤微环境代谢重编程等分子机制等[17-18],可以为合理的科学假设的提出奠定坚实的基础,这将为胰腺癌的临床与基础研究提供新的途径。
TIMER数据库能够从RNA-seq表达谱数据中,检测和量化肿瘤组织中免疫细胞浸润的情况,由此来确定肿瘤细胞-免疫细胞之间的关系。利用TIMER数据库可以精准量化肿瘤纯度、免疫细胞浸润的丰度,发现与患者临床预后信息的相关性。本研究通过TIMER数据库分析发现,在胰腺癌局部免疫微环境中,EGFR和LDHA mRNA表达呈显著性正相关。研究[19]表明EGFR基因拷贝数增加或者过度表达均能促进正常细胞转化、维持肿瘤干性特征和肿瘤转移。Chen等[20]发现,在KRAS致癌基因突变发生,EGFR诱导表达NFATc1和SOX9基因,导致腺泡细胞转分化和引发胰腺癌。Shi等[21]发现,LDHA和转录因子Krüppel样因子4(KLF4)在胰腺肿瘤组织中表达呈负相关;在转录水平,KLF4作用LDHA启动子区域,负调控LDHA基因转录活性,抑制LDHA促进的胰腺癌糖酵解。He等[22]也发现LDHA通过c-Myc信号通路调控胰腺癌糖酵解。业已证实,肿瘤细胞代谢和局部免疫微环境抑制密切相关[23]。本研究利用相关性分析方法,发现EGFR和LDHA的表达与EGFR、LDHA、P38、FOXM1和CSF1R等表达相关,推测EGFR和LDHA可能是通过这些信号分子参与局部免疫微环境细胞侵袭转移影响生存。
胰腺癌恶性程度极高,具有较强的肿瘤异质性,对其进行精准的分期是指导临床治疗和监测预后具有重要意义。美国癌症联合委员会(AJCC)推出的TNM分期系统已成为当前肿瘤分期的“金标准”,并不断得以更新。日前,AJCC基于最新的循证医学证据发布了第8版胰腺癌分期系统,强调肿瘤大小是影响患者的生存独立高危因素[24]。尤为重要的是,第8版胰腺癌分期系统相较第7版分期更细与预后的相关性更大;定义更为清晰,判断标准更突出客观性的可测量指标,摒除了主观性的指标。T1~T3根据肿瘤大小界定,不再使用肿瘤胰腺外侵犯的概念;T4是指肿瘤侵犯腹腔干动脉、肠系膜上动脉和(或)肝总动脉,摒弃可切除性的定义[25]。本研究显示EGFR和LDHA基因mRNA表达和预后生存之间具有显著的相关性,高表达患者预后差。通过Cox比例风险回归模型分析发现,LDHA较EGFR能更为准确的预测胰腺癌的临床预后。提示LDHA基因在胰腺癌中有可能作为临床预后指标。但是结果显示胰腺癌预后与TNM分期无关,笔者深入分析发现TIMER数据库纳入的胰腺癌样本分期标准是基于AJCC第6版标准,此外TIMER数据库是基于把TNM分期作为协变量,通过拟合函数方法在线分析,可能导致Cox比例风险回归模型分析结果无统计学意义。EGFR和LDHA基因表达联合新的TNM分期预测预后还有待在新的队列样本中验证。
尽管目前多数研究提示,EDFR和LDHA确实参与了多种肿瘤的发生发展过程,是一种重要的信号分子,但在胰腺癌局部免疫微环境中发挥了何种作用及如何调控,还需通过体外细胞实验和体内动物模型实验更深入探讨EGFR和LDHA的作用及机制,为胰腺癌的免疫治疗提供新思路。
[1] Kamisawa T, Wood LD, Itoi T, et al. Pancreatic cancer[J]. Lancet,2016, 388(10039):73–85. doi: 10.1016/S0140–6736(16)00141–0.
[2] 杨永超, 李宜雄. 胰腺癌外科治疗的历史和现状[J]. 中国普通外科杂志, 2018, 27(3):269–283. doi:10.3978/j.issn.1005–6947.2018.03.002.Yang YC, Li YX. The surgical treatment of pancreatic cancer:history and present state[J]. Chinese Journal of General Surgery,2018, 27(3):269–283. doi:10.3978/j.issn.1005–6947.2018.03.002.
[3] Qian ZR, Rubinson DA, Nowak JA, et al. Association of Alterations in Main Driver Genes With Outcomes of Patients With Resected Pancreatic Ductal Adenocarcinoma[J]. JAMA Oncol, 2018,4(3):e173420. doi: 10.1001/jamaoncol.2017.3420.
[4] Mondal G, Kumar V, Shukla SK, et al. EGFR-targeted polymeric mixed micelles carrying gemcitabine for treating pancreatic cancer[J]. Biomacromolecules, 2016, 17(1):301–313. doi: 10.1021/acs.biomac.5b01419.
[5] Brand A, Singer K, Koehl GE, et al. LDHA-Associated Lactic Acid Production Blunts Tumor Immunosurveillance by T and NK Cells[J]. Cell Metab, 2016, 24(5):657–671. doi: 10.1016/j.cmet.2016.08.011.
[6] Oettle H, Lehmann T. Gemcitabine-resistant pancreatic cancer:a second-line option[J]. Lancet, 2016, 387(10018):507–508. doi:10.1016/S0140–6736(15)01035–1.
[7] Chong CR, Jänne PA. The quest to overcome resistance to EGFR-targeted therapies in cancer[J]. Nat Med, 2013, 19(11):1389–1400.doi: 10.1038/nm.3388.
[8] Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells[J].Cancer Res, 2017, 77(21):e108-e110. doi: 10.1158/0008–5472.CAN-17–0307.
[9] Alam MS, Gaida MM, Bergmann F, et al. Selective inhibition of the p38 alternative activation pathway in infiltrating T cells inhibits pancreatic cancer progression[J]. Nat Med, 2015, 21(11):1337–1343. doi: 10.1038/nm.3957.
[10] Cui J, Shi M, Xie D, et al. FOXM1 promotes the warburg effect and pancreatic cancer progression via transactivation of LDHA expression[J] Clin Cancer Res, 2014, 20(10):2595–2606. doi:10.1158/1078–0432.CCR-13–2407.
[11] Mace TA, Shakya R, Pitarresi JR, et al. IL-6 and PD-L1 antibody blockade combination therapy reduces tumour progression in murine models of pancreatic cancer[J]. Gut, 2018, 67(2):320–332.doi: 10.1136/gutjnl-2016–311585.
[12] Zhu Y, Knolhoff BL, Meyer MA, et al. CSF1/CSF1R blockade reprograms tumor‐infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models[J].Cancer Res, 2014, 74(18):5057–5069. doi: 10.1158/0008–5472.CAN-13–3723.
[13] Wu C, Kraft P, Stolzenberg-Solomon R, et al. Genomewide association study of survival in patients with pancreatic adenocarcinoma[J]. Gut, 2014, 63(1):152–160. doi: 10.1136/gutjnl-2012–303477.
[14] Rizzato C, Campa D, Talar-Wojnarowska R, et al. Association of genetic polymorphisms with survival of pancreatic ductal adenocarcinoma patients[J]. Carcinogenesis, 2016, 37(10):957–964.doi: 10.1093/carcin/bgw080.
[15] Frampton AE, Castellano L, Colombo T, et al. Integrated molecular analysis to investigate the role of microRNAs in pancreatic tumour growth and progression[J]. Lancet, 2015, 385(Suppl 1): S37. doi:10.1016/S0140–6736(15)60352-X.
[16] 张志鹏, 陆晔斌, 陈泓西, 等. 基于生物信息学方法的胰腺导管腺癌预后风险标志物筛选[J]. 中国普通外科杂志, 2017,26(9):1097–1104. doi:10.3978/j.issn.1005–6947.2017.09.004.Zhang ZP, Lu YB, Chen HX, et al. Screening of prognostic risk markers for pancreatic ductal adenocarcinoma based on bioinformatics approaches[J]. Chinese Journal of General Surgery,2017, 26(9):1097–1104. doi:10.3978/j.issn.1005–6947.2017.09.004.
[17] Makohon-Moore AP, Zhang M, Reiter JG, et al. Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer[J]. Nat Genet, 2017,49(3):358–366. doi: 10.1038/ng.3764.
[18] McDonald OG, Li X, Saunders T, et al. Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis[J]. Nat Genet, 2017, 49(3):367–376. doi: 10.1038/ng.3753.
[19] Seguin L, Kato S, Franovic A, et al. An integrin β3-KRASRalB complex drives tumour stemness and resistance to EGFR inhibition[J]. Nat Cell Biol, 2014, 16(5):457–468. doi: 10.1038/ncb2953.
[20] Chen NM, Singh G, Koenig A, et al. NFATc1 Links EGFR Signaling to Induction of Sox9 Transcription and Acinar–Ductal Transdifferentiation in the Pancreas[J]. Gastroenterology, 2015,148(5):1024–1034. doi: 10.1053/j.gastro.2015.01.033.
[21] Shi M, Cui J, Du J, et al. A novel KLF4/LDHA signaling pathway regulates aerobic glycolysis in and progression of pancreatic cancer[J]. Clin Cancer Res, 2014, 20(16):4370–4380. doi:10.1158/1078–0432.CCR-14–0186.
[22] He TL, Zhang YJ, Jiang H, et al. The c-Myc–LDHA axis positively regulates aerobic glycolysis and promotes tumor progression in pancreatic cancer[J]. Med Oncol, 2015, 32(7):187. doi: 10.1007/s12032–015–0633–8.
[23] Quaranta V, Rainer C, Nielsen SR, et al. Macrophage-derived granulin drives resistance to immune checkpoint inhibition in metastatic pancreatic cancer[J]. Cancer Res, 2018, 78(15):4253–4269. doi: 10.1158/0008–5472.CAN-17–3876.
[24] Yu J, Blackford AL, Dal Molin M, et al. Time to progression of pancreatic ductal adenocarcinoma from low-to-high tumour stages[J]. Gut, 2015, 64(11):1783–1789. doi: 10.1136/gutjnl-2014–308653.
[25] 白雪莉, 马涛, 梁廷波. 美国癌症联合委员会第8版胰腺癌分期系统更新简介及解读[J]. 中国实用外科杂志, 2017, 37(2):146–148.doi: 10.19538/j.cjps.issn1005–2208.2017.02.11.Bai XL, Ma T, Liang TB. Update of the 8th edition of AJCC cancer staging for pancreatic cancer[J]. Chinese Journal of Practical Surgery, 2017, 37(2):146–148. doi: 10.19538/j.cjps.issn1005–2208.2017.02.11.
