原发性肝癌(primary hepatocellular carcinoma,PHC)是指由肝细胞或肝内胆管上皮细胞发生的恶性肿瘤,其中肝细胞癌占85%以上,是我国第四位常见的恶性肿瘤,病死率位居第三[1]。肝细胞癌恶性程度高,易发生转移,许多患者确诊时已发展为晚期,5年生存率小于5%[2]。驱动蛋白超家族(kinesin superfamily proteins,KIFs)是一类分子马达,通过水解ATP产生能量来进行细胞内囊泡、细胞器、RNA等物质的运输[3-4]。驱动蛋白家族成员15(kinesin family member 15,KIF15)是该家族中的一员,属于kinesin-12亚家族,又称hklp2,是除Kinesin-5之外第二个四聚体纺锤体马达[5-6]。已有研究[7-9]表明,KIF与乳腺癌、肺癌、胰腺癌的发生发展过程相关,然而其在肝癌中的研究尚未报道。本研究通过生物信息学分析KIF15与肝细胞癌的关系,并通过下调其表达探究其对肝细胞癌转移侵袭能力的影响。
1 材料与方法
1.1 细胞株与主要试剂
人肝癌细胞系SMMC-7721由华中科技大学附属同济医院胆胰外科实验室馈赠。DMEM培养基、胎牛血清购自美国Gibco公司,siKIF15购自广州锐博生物科技有限公司,Lipofectamine2000购自美国赛默飞公司,Transwell小室及六孔板购自美国Corning公司,KIF15、GAPDH抗体购自美国CST公司,CCK-8试剂购自日本Dojindo公司,Matrigel基质胶购于美国BD公司。
1.2 实验方法
1.2.1 细胞培养 人肝癌细胞系SMMC-7721在含10%胎牛血清的DMEM培养基、37 ℃、5%CO2的条件下,于细胞培养箱中培养,根据细胞生长情况每2~3天更换新鲜培养基或进行细胞传代培养。
1.2.2 生物信息学分析 从UCSC Xena网站下载TCGA Liver Cancer肝癌数据中KIF15的表达情况及相关临床资料[10]。基因表达使用标准化高敏荧光序列进行分析,将标准化计数结果作为基因表达程度的估计值。分析得到不同TNM分期肝癌患者体内KIF15的表达以及肝癌患者的生存曲线。
1.2.3 siRNA转染 选取处于对数生长期的SMMC-7721细胞制备细胞悬液,取2×105个细胞接种于六孔板中,待细胞贴壁后按照lipofectamine 2000试剂说明书将siKIF15及siControl分别转染入细胞中作为实验组及对照组。Western blot检测实验组及对照组细胞中KIF15表达量。
1.2.4 CCK-8法检测细胞增殖活性 分别取转染了siKIF15的实验组及转染了siControl的对照组SMMC-7721细胞,调整细胞密度为2×104个/mL接种于96孔培养板中,每孔100 μL,置于培养箱中培养。待细胞完全贴壁后吸去培养液,于贴壁0、24、48、72 h后每孔加入20 μL CCK-8,继续培养4 h,于酶标仪波长450 nm处读取吸光度(OD)值。
1.2.5 流式细胞术检测细胞周期 分别收集实验组及对照组细胞,PBS洗涤后缓慢滴入预冷的无水乙醇,调整乙醇浓度至75%,充分振荡,-20 ℃固定过夜。800 r/min,5 min收集细胞,PBS洗涤后加入 50 μL RNase及 50 μL碘化丙啶(PI),室温避光孵育30 min。流式细胞仪检测细胞周期分布,采用CellQuest及ModFit软件分析实验结果。
1.2.6 Trasnwell侵袭实验 Matrigel基质胶用无血清培养基按1:8稀释,加入小室上室面,4 ℃过夜。第2天于37 ℃ 30 min水化基底膜,同时制备转染48 h后的SMMC-7721细胞悬液(用不含血清的培养基重悬),密度为2×105个/mL,每个小室中加入200 μL,下室加入含10%血清的培养基600 μL,置于培养箱中培养24~36 h后用PBS洗两遍,4%多聚甲醛固定30 min,0.1%结晶紫染色30 min,用棉签擦去上室面细胞,晾干并拍照。
1.3 统计学处理
采用SPSS 23.0对实验数据进行分析、整理。计量资料采用均数±标准差(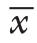 ±s)表示,两组比较采用t检验,P<0.05为差异有统计学意义。
±s)表示,两组比较采用t检验,P<0.05为差异有统计学意义。
2 结 果
2.1 KIF15在不同临床资料分组的肝癌患者中的相对表达情况
通过分析得出,T2~T4分期的肝癌患者中K I F 15相对表达量明显高于T1分期的患者[(-1.337±2.249) vs. (-1.863±1.924);t=2.562,P<0.05];TNM分期在II~IV期的患者KIF15相对表达量明显高于TNM分期为I期的患者[(-1.282±2.229) vs. ( -1.839±1.921);t=2.639,P<0.01](图1)。KIF15表达量在淋巴结转移阳性组和淋巴结转移阴性无统计学差异(P>0.05)。
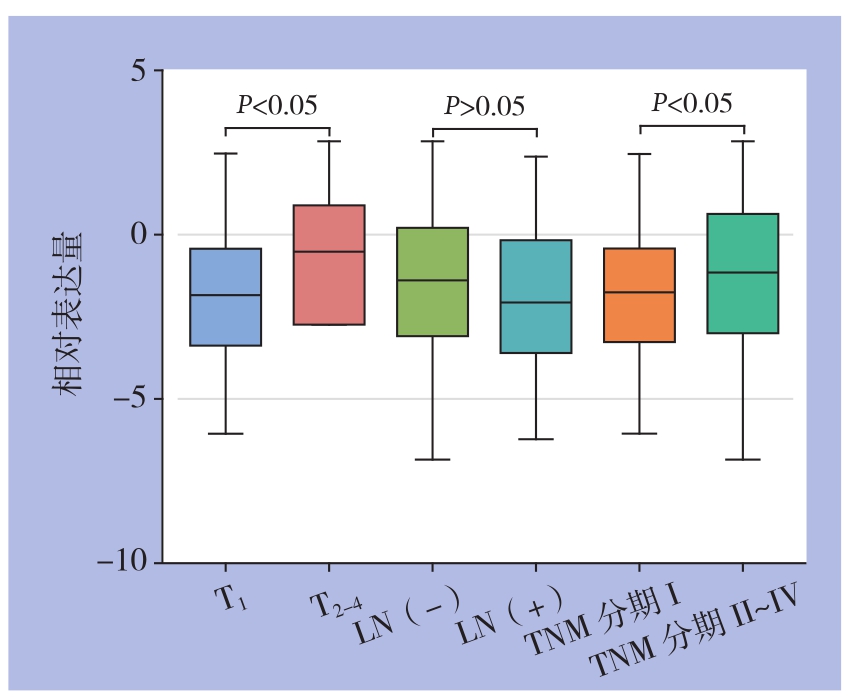
图1 KIF15在不同临床资料分组的肝癌患者中的表达比较
Figure1 Comparison of KIF15 expressions between patients grouped by different clinical data
2.2 KIF15表达量对肝癌患者生存率的影响
通过绘制生存曲线并进行统计学分析得出,KIF15相对表达量≥-1.45的肝癌患者生存率明显低于KIF15相对表达量<-1.45的患者,差异具有统计学意义(P<0.05)(图2)。
2.3 Western blot检测KIF15沉默效率
将构建好的siKIF15转染入SMMC-7721细胞系中,Western blot检测转染效率,结果表明,转染了siKIF15的细胞KIF15蛋白表达量明显低于转染siControl的对照组(图3)。
2.4 沉默KIF15抑制了SMMC-7721的增殖
CCK-8结果表明,转染了siKIF15的实验组SMMC-7721细胞增殖活性较对照组细胞有所降低。其中,实验组细胞在96 h的吸光度为1.826±0.079,对照组为2.374±0.156,差异有统计学意义(t=3.124,P<0.05)(图4)。

图2 不同KIF15表达量肝癌患者的生存曲线
Figure2 Survival curves of liver cancer patients with different KIF15 expressions
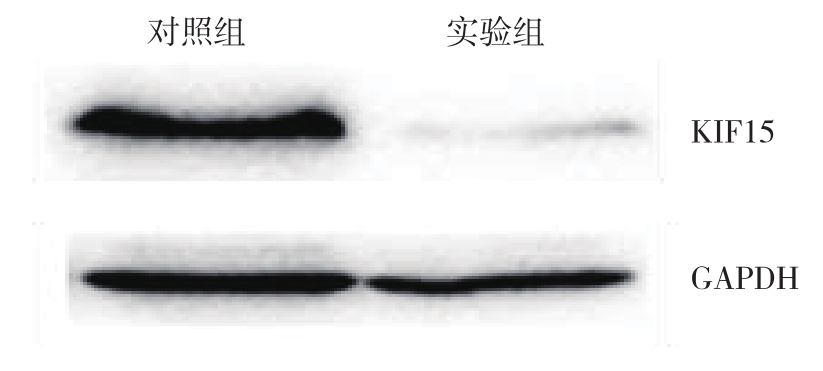
图3 Western blot检测沉默效率
Figure3 Silencing efficiency detection by Western blot
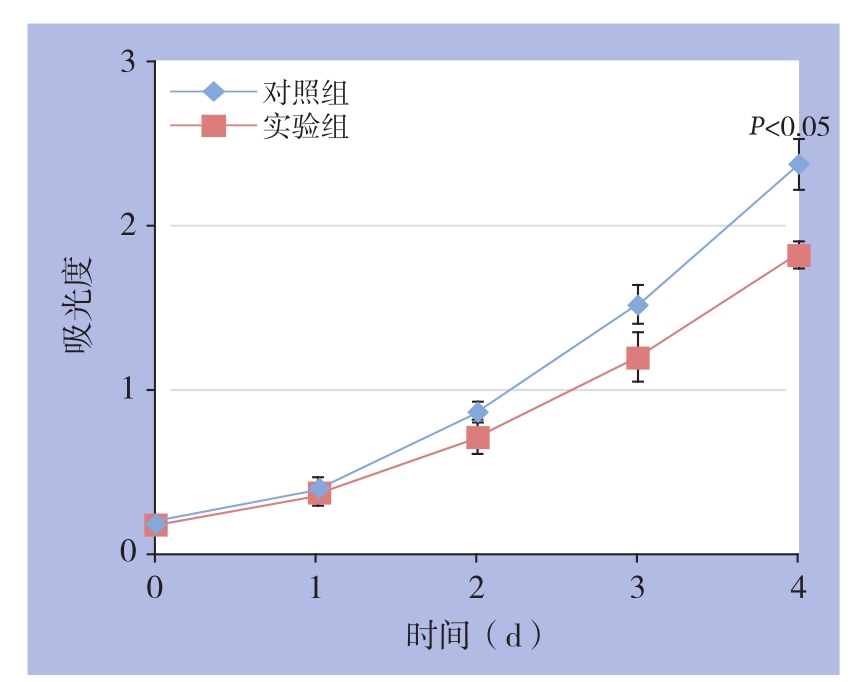
图4 细胞增殖曲线
Figure4 Cell proliferation curves
2.5 沉默KIF15使SMMC-7721阻滞在G0/G1期
流式细胞术结果表明,转染了siKIF 15的实验组SMMC-7721细胞G1期比例高于对照组,差异具有统计学意义[(81.04±2.47)% vs.(64.98%±2.67)%,t=4.417,P<0.05];S期比例低于对照组,差异具有统计学意义[(13.35±2.16)% vs.(28.48±2.14)%,t=4.978,P<0.01];G2期比例无统计学差异(P>0.05)(图5)。
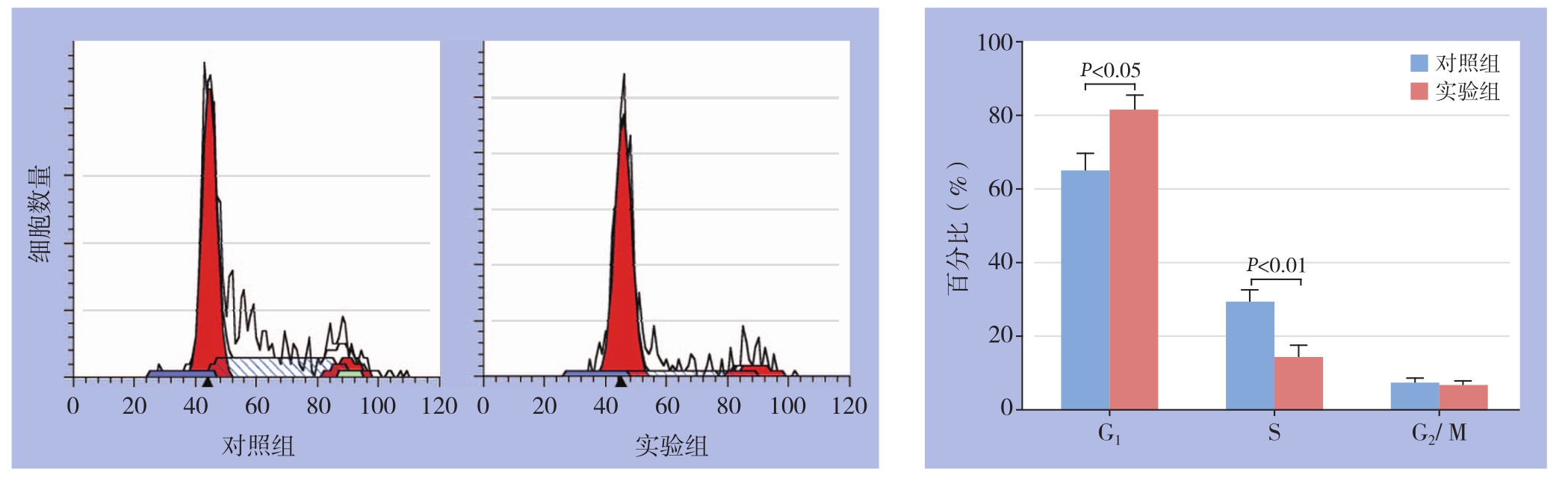
图5 细胞周期分析
Figure5 cell cycle analysis
2.6 沉默KIF15降低了SMMC-7721的侵袭能力
Transwell实验结果表明,转染了siKIF15的SMMC-7721细胞侵袭数目为(417±164)个,明显低于对照组细胞侵袭数目(951±159)个,差异有统计学意义(t=4.679,P<0.01)(图6)。

图6 细胞侵袭力检测
Figure6 Determination of invasion ability of the cells
3 讨 论
近年来大量研究[11-13]表明,KIFs是一类保守的微管依赖的分子运动蛋白,具有ATP酶活性和运动特征,其与神经退行性疾病、代谢性疾病、肾病等多种疾病均有关联。除此之外,KIFs在肿瘤的发生发展过程中也起到重要作用[14-17]。学者[18-20]认为,在肿瘤细胞进行分裂的过程中,异常表达的KIFs将通过影响染色体的凝集、纺锤体的形成发生异常,进而影响肿瘤细胞增殖和侵袭转移能力。例如在神经胶质瘤中,KIF1B通过诱导膜型基质金属蛋白酶促进胶质瘤细胞的迁移和侵袭[21]。在沉默KIF2A后,乳腺癌细胞的增殖和迁移能力被明显抑制,并且其与乳腺癌的预后密切相关[22]。而在肺癌细胞中,下调KIF23促进凋亡的发生,其被认为是肺癌患者潜在治疗靶点[23]。肝细胞癌是一类恶性程度极高的肿瘤,KIFs对肝癌发生发展的作用尚不明确[24]。本研究着重分析TCGA数据库中KIF15的表达与临床资料的相关性,探究KIF15对肝癌恶性生物学行为的影响。
本研究首先通过TCGA Liver Cancer肝癌数据分析得出KIF15在TNM II~IV期患者的表达量明显高于TNM I期患者,其中T分期为2~4期的患者KIF15表达量明显高于T1期患者。而进一步通过绘制生存曲线并统计分析得出KIF15高表达的肝癌患者生存率明显低于KIF15低表达患者,提示KIF15可以作为判断肝癌预后的指标,即KIF15高表达患者临床预后较差。在细胞实验中,将siKIF15导入肝癌细胞并通过Western blot验证了其沉默效率。CCK-8实验表明沉默KIF15后肝癌细胞增殖能力明显降低,进一步分析细胞周期发现沉默KIF15能够将肝癌细胞周期阻滞在G0/G1期,表明KIF15可以通过促进肝癌细胞的G1/S期转化而加速其增殖。接下来本研究还探讨了KIF15与肝癌细胞侵袭能力的关系,Transwell实验表明,相比于对照组,沉默KIF15肝癌细胞侵袭能力明显下降,表明KIF15在肝癌细胞的侵袭过程中发挥了一定的促进作用。在Eskova等[4]的研究中,KIF15介导网格蛋白Dab2在细胞内的定位,调节整合素的内化过程从而影响了细胞的侵袭能力。Wang等[25]也发现KIF15通过激活MEK-ERK通路加速了细胞的G1/S期转化,促进了胰腺癌细胞的异常增殖。以上研究结果均提示KIF15在扮演了促进肿瘤进展的角色。
综上所述,KIF15在肝癌的增殖和侵袭过程中发挥了一定的作用,并可以作为判断肝癌患者预后的指标,有望成为肝癌早期诊断的标志物及化疗的潜在靶点,但其促进肝癌细胞侵袭转移的具体机制还需进一步深入研究。
[1] 中华人民共和国国家卫生和计划生育委员会. 原发性肝癌诊疗规范(2017年版)[J]. 临床肝胆病杂志, 2017, 33(8):1419-1431.doi:10.3969/j.issn.1001-5256.2017.08.003.Bureau of Medical Administration, National Health and Family Planning Commission. Diagnosis, management, and treatment of hepatocellular carcinoma (V2017)[J]. Journal of Clinical Hepatology, 2017, 33(8):1419-1431. doi:10.3969/j.issn.1001-5256.2017.08.003.
[2] Dutta R, Mahato RI. Recent advances in hepatocellular carcinoma therapy[J]. Pharmacol Ther, 2017, 173:106-117. doi:10.1016/j.pharmthera.2017.02.010.
[3] Liu M, Nadar VC, Kozielski F, et al. Kinesin-12, a mitotic microtubule-associated motor protein, impacts axonal growth,navigation, and branching[J]. J Neurosci, 2010, 30(44):14896-14906. doi:10.1523/JNEUROSCI.3739-10.2010.
[4] Eskova A, Knapp B, Matelska D, et al. An RNAi screen identifies KIF15 as a novel regulator of the endocytic trafficking of integrin[J]. J Cell Sci, 2014, 127(Pt 11):2433-2447. doi:10.1242/jcs.137281.
[5] Florian S, Mayer TU. Modulated microtubule dynamics enable Hklp2/Kif15 to assemble bipolar spindles[J]. Cell Cycle, 2011,10(20):3533-3544. doi:10.4161/cc.10.20.17817.
[6] Sebastian J. Dihydropyrazole and dihydropyrrole structures based design of Kif15 inhibitors as novel therapeutic agents for cancer[J]. Comput Biol Chem, 2017, 68:164-174. doi:10.1016/j.compbiolchem.2017.03.006.
[7] Zou JX, Duan Z, Wang J, et al. Kinesin Family Deregulation Coordinated by Bromodomain Protein ANCCA and Histone Methyltransferase MLL for Breast Cancer Cell Growth, Survival,and Tamoxifen Resistance[J]. Mol Cancer Res, 2014, 12(4):539-549. doi:10.1158/1541-7786.MCR-13-0459.
[8] Bidkhori G, Narimani Z, Ashtiani SH, et al. Reconstruction of an integrated genome-scale co-expression network reveals key modules involved in lung adenocarcinoma[J]. Plos One, 2013,8(7):e67552. doi:10.1371/journal.pone.0067552.
[9] Yokota K, Sasaki H, Okuda K, et al. KIF5B/RET fusion gene in surgically-treated adenocarcinoma of the lung[J]. Oncol Rep, 2012,28(4):1187-1192. doi:10.3892/or.2012.1908.
[10] TCGA. Liver Cancer (LIHC) dataset. UCSC Xena, USA[DB].https://xenabrowser.net/. Accessed 16 Jan 2018.
[11] Yang W, Tanaka Y, Bundo M, et al. Antioxidant signaling involving the microtubule motor KIF12 is an intracellular target of nutrition excess in beta cells[J]. Dev Cell, 2014, 31(2):202-214. doi:10.1016/j.devcel.2014.08.028.
[12] Lin F, Hiesberger T, Cordes K, et al. Kidney-specific inactivation of the KIF3A subunit of kinesin-II inhibits renal ciliogenesis and produces polycystic kidney disease[J]. Proc Natl Acad Sci U S A,2003, 100(9):5286-5291. doi:10.1073/pnas.0836980100.
[13] Inomata H, Nakamura Y, Hayakawa A, et al. A scaffold protein JIP-1b enhances amyloid precursor protein phosphorylation by JNK and its association with kinesin light chain 1[J]. J Biol Chem, 2003,278(25):22946-22955. doi:10.1074/jbc.M212160200.
[14] Chen J, Li S, Zhou S, et al. Kinesin superfamily protein expression and its association with progression and prognosis in hepatocellular carcinoma[J]. J Cancer Res Ther, 2017, 13(4):651-659. doi:10.4103/jcrt.JCRT_491_17.
[15] Venere M, Horbinski C, Crish JF, et al. The mitotic kinesin KIF11 is a driver of invasion, proliferation, and self-renewal in glioblastoma[J]. Sci Transl Med, 2015, 7(304):304ra143. doi:10.1126/scitranslmed.aac6762.
[16] Shichijo S, Ito M, Azuma K, et al. A unique gene having homology with the kinesin family member 18A encodes a tumour-associated antigen recognised by cytotoxic T lymphocytes from HLAA2+colon cancer patients[J]. Eur J Cancer, 2005, 41(9):1323-1330.doi:10.1016/j.ejca.2005.02.025
[17] Sun XT, Jin ZT, Song X, et al. Evaluation of KIF23 variant 1 expression and relevance as a novel prognostic factor in patients with hepatocellular carcinoma[J]. BMC Cancer, 2015, 15:961. doi:10.1186/s12885-015-1987-1.
[18] Mazumdar M, Lee JH, Sengupta K, et al. Tumor formation via loss of a molecular motor protein[J]. Curr Biol, 2006, 16(15):1559-1564. doi:10.1016/j.cub.2006.06.029.
[19] Myers SM, Collins I. Recent findings and future directions for interpolar mitotic kinesin inhibitors in cancer therapy[J]. Future Med Chem, 2016, 8(4):463-489. doi:10.4155/fmc.16.5.
[20] Yu Y, Feng YM. The role of kinesin family proteins in tumorigenesis and progression:potential biomarkers and molecular targets for cancer therapy[J]. Cancer, 2010, 116(22):5150-5160. doi:10.1002/cncr.25461.
[21] Chen S, Han M, Chen W, et al. KIF1B promotes glioma migration and invasion via cell surface localization of MT1-MMP[J]. Oncol Rep, 2016, 35(2):971-977. doi:10.3892/or.2015.4426.
[22] Wang J, Ma S, Ma R, et al. KIF2A silencing inhibits the proliferation and migration of breast cancer cells and correlates with unfavorable prognosis in breast cancer[J]. BMC Cancer, 2014,14:461. doi:10.1186/1471-2407-14-461.
[23] Iltzsche F, Simon K, Stopp S, et al. An important role for Myb-MuvB and its target gene KIF23 in a mouse model of lung adenocarcinoma[J]. Oncogene, 2017, 36(1):110-121. doi:10.1038/onc.2016.181.
[24] Sia D, Villanueva A, Friedman SL, et al. Liver Cancer Cell of Origin, Molecular Class, and Effects on Patient Prognosis[J].Gastroenterology, 2017, 152(4):745-761. doi:10.1053/j.gastro.2016.11.048.
[25] Wang J, Guo X, Xie C, et al. KIF15 promotes pancreatic cancer proliferation via the MEK-ERK signalling pathway[J]. Br J Cancer,2017, 117(2):245-255. doi:10.1038/bjc.2017.165.
