肝细胞癌(hepatocellular carcinoma,HCC)恶性程度高,其发病率在所有肿瘤中高居第4位,致死率较高[1-3],中国每年HCC死亡例数占约占全球总数的55%,现已成为严重威胁我国人民群众健康的重大恶性肿瘤之一[4-5]。目前临床医生对于HCC的诊断工具主要依靠彩超、CT、MR及AFP等检查、检验,但HCC起病隐匿,症状不明显,待发现时大多已属于中晚期,手术切除、射频消融、介入等治疗方法有效的提高了HCC患者生存率[6],但总体预后难以令人满意,5年生存率约30%~40%[7]。因此,寻找新的HCC诊断分子标志物意义仍然重大。
目前,高通量测序技术能够从整个基因组或转录组水平探索疾病的发生、发展,已广泛用于一些疾病基因表达谱分析、基因克隆和寻找特异性分子标志物[8-9]。白文萱等[10]通过生物学信息分析的方法,对HCC基因芯片数据进行挖掘,发现磷脂酰肌醇蛋白聚糖(GPC3)在HCC中表达上调,可作为HCC特异性的免疫治疗靶点;Zhang等[11]通过高通量测序生物学分析的方法发现HCC差异表达的基因在抗原处理与呈递、补体级联和 I型干扰素信号通路有协同或拮抗作用,而这些对HCC的发生与侵袭能力变化有重要影响,高通量测序技术是测序技术发展的里程碑,它为现代生命技术研究提供了前所未有的机遇[12]。
外泌体是细胞分泌的功能性小体,直径为30~150 nm,在人体血液、唾液、尿液等体液中均能够被检测到[13]。其体积微小并且自身稳定,脂质双层膜结构可有效保护miRNA、mRNA和其它非编码RNA核酸分子不被降解[14-15],被认为是肿瘤标志物的天然载体,近年在肿瘤诊断、发生发展机制、靶向治疗中越来越得到重视。Wang等[16]发现外泌体miR-122、miR-1246和miR-148a在HCC患者中呈显著性高表达,其中miR-148a诊断效能显著优于AFP(0.891 vs.0.712),此外miR-122、AFP和miR-148a联合用于区分HCC和肝硬化患者的诊断效能高达0.931。有学者发现HCC患者血清外泌体中的miR-9-3P水平明显降低,成纤维细胞因子5(HBGF-5)在细胞增殖中具有重要作用,后者是miR-9-3P潜在的靶mRNA,miR-9-3P过表达显著下调了HBGF-5在mRNA和蛋白水平的表达,进而影响了HCC细胞的增殖[17]。
综上,笔者推测HCC患者与正常人之间血清外泌体mRNA存在较大差异,可能筛选出有意义的分子标志物,故拟通过高通量测序方法进行检测,以期寻找合适的HCC诊断标志物。
1 资料与方法
1.1 一般资料
收集2018年8月—2018年12月在新乡医学院第一附属医院就诊的HCC患者血液标本3例(Ca1、Ca2、Ca3),均有乙型肝炎及肝硬化病史,且肿瘤病理类型均经病理科医师确诊 (其中Ca1为中分化HCC,部分侵及肝被膜,可见脉管内癌栓;Ca2为低分化HCC,紧邻被膜;Ca3为中分化HCC)。3例HCC患者中男2例、女1例。同时收集3例正常志愿者血液标本(N1、N2、N3)作对照,3例正常志愿者中男2例、女1例。采集样本前均未接受临床治疗。本研究经医院伦理委员会批准,且研究对象均知情同意并签署知情同意书。
1.2 仪器和主要试剂
仪器:illumina Hiseq Xten;模式:PE150;试剂:Agencourt Ampure XP beads,美国Beckman Coulter公司;NEBNext® Ultra™ RNA Library Prep Kit for Illumina,美国NEB公司;HiSeq Rapid SBS Kit V2(200 cycle),美国Illumina公司;HiSeq Rapid PE Cluster Kit V2,美国Illumina公司。
1.3 血清外泌体的提取及鉴定
所有研究对象采集静脉血6 mL,EDTA-K2抗凝管收集静置,然后离心取血清。血清外泌体采用锐博外泌体提取试剂盒RiboTM Exosome Isolation Reagent(for plasma or serum)提取,严格按照试剂说明进行操作。外泌体表面蛋白标志物采用Western blot进行检测,所用抗体包括CD9、CD63、TSGl01及羊抗兔二抗。
1.4 外泌体RNA 提取
用Magen试剂盒提取外泌体RNA,操作步骤详见说明书。
1.5 外泌体mRNA 高通量测序
首先对HCC患者血清外泌体mRNA进行纯化等前期处理,然后进行反转录形成cDNA、PCR扩增等实验操作,再经过文库质检后上机测序;再对测序质量进行评价,以取得高质量的数据,并对高质量的数据及参考数据进行比对,获得BAM文件。上机测序由广州市锐博生物公司测序,并进行差异表达外泌体mRNA进行分析,采用AUdics对差异表达mRNA进行分析。对差异表达的外泌体mRNA进行GO(Gene Ontology)和KEGG pathway分析。
2 结 果
2.1 差异外泌体mRNA 筛选
筛选的差异表达外泌体mRNA统计结果如 图1,其中HCC组相对正常人组上调397个外泌体mRNA,192个外泌体mRNA表达下调。

图1 mRNA 差异表达火山图
Figure1 Volcanic map of the mRNAs with differential expression
2.2 显著差异表达的外泌体mRNA
对差异外泌体mRNA进行表达差异性分析,采用HCC组相对正常人组外泌体mRNA表达fold change(log2,表达差异倍数)对差异蛋白进行筛选,挑选条件如下:Log2≤1.9或者Log2≥2.5;表达水平显著增加的外泌体mRNA17个,表达水平明显降低的外泌体mRNA14个(表1)。
表1 两组表达差异显著的外泌体mRNA
Table1 The exosomal mRNAs with significantly different expressions between the two group
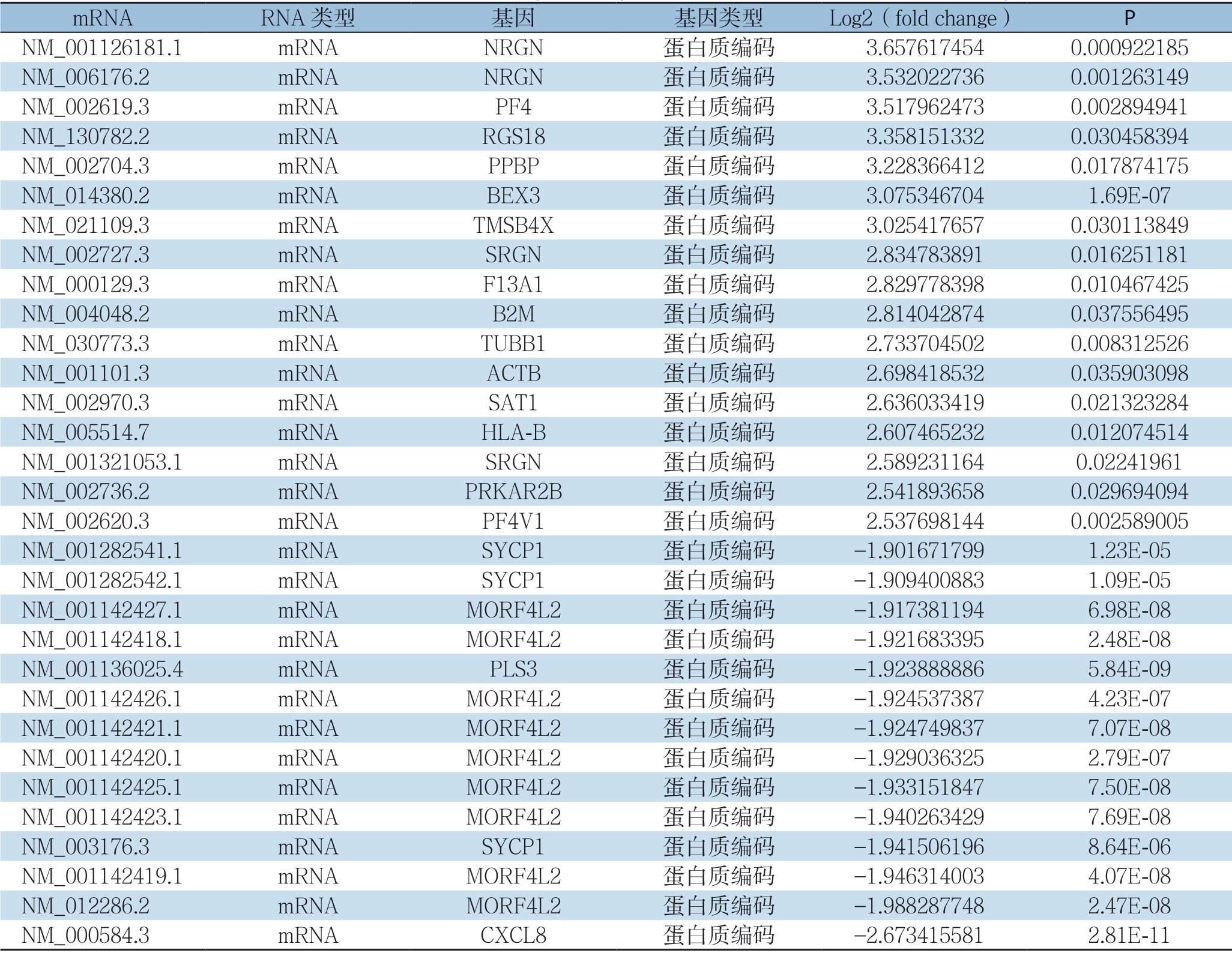
mRNA RNA 类型 基因 基因类型 Log2(fold change) P NM_001126181.1 mRNA NRGN 蛋白质编码 3.6576174540.000922185 NM_006176.2 mRNA NRGN 蛋白质编码 3.5320227360.001263149 NM_002619.3 mRNA PF4 蛋白质编码 3.5179624730.002894941 NM_130782.2 mRNA RGS18 蛋白质编码 3.3581513320.030458394 NM_002704.3 mRNA PPBP 蛋白质编码 3.2283664120.017874175 NM_014380.2 mRNA BEX3 蛋白质编码 3.0753467041.69E-07 NM_021109.3 mRNA TMSB4X 蛋白质编码 3.0254176570.030113849 NM_002727.3 mRNA SRGN 蛋白质编码 2.8347838910.016251181 NM_000129.3 mRNA F13A1 蛋白质编码 2.8297783980.010467425 NM_004048.2 mRNA B2M 蛋白质编码 2.8140428740.037556495 NM_030773.3 mRNA TUBB1 蛋白质编码 2.7337045020.008312526 NM_001101.3 mRNA ACTB 蛋白质编码 2.6984185320.035903098 NM_002970.3 mRNA SAT1 蛋白质编码 2.6360334190.021323284 NM_005514.7 mRNA HLA-B 蛋白质编码 2.6074652320.012074514 NM_001321053.1 mRNA SRGN 蛋白质编码 2.5892311640.02241961 NM_002736.2 mRNA PRKAR2B 蛋白质编码 2.5418936580.029694094 NM_002620.3 mRNA PF4V1 蛋白质编码 2.5376981440.002589005 NM_001282541.1 mRNA SYCP1 蛋白质编码 -1.9016717991.23E-05 NM_001282542.1 mRNA SYCP1 蛋白质编码 -1.9094008831.09E-05 NM_001142427.1 mRNA MORF4L2 蛋白质编码 -1.9173811946.98E-08 NM_001142418.1 mRNA MORF4L2 蛋白质编码 -1.9216833952.48E-08 NM_001136025.4 mRNA PLS3 蛋白质编码 -1.9238888865.84E-09 NM_001142426.1 mRNA MORF4L2 蛋白质编码 -1.9245373874.23E-07 NM_001142421.1 mRNA MORF4L2 蛋白质编码 -1.9247498377.07E-08 NM_001142420.1 mRNA MORF4L2 蛋白质编码 -1.9290363252.79E-07 NM_001142425.1 mRNA MORF4L2 蛋白质编码 -1.9331518477.50E-08 NM_001142423.1 mRNA MORF4L2 蛋白质编码 -1.9402634297.69E-08 NM_003176.3 mRNA SYCP1 蛋白质编码 -1.9415061968.64E-06 NM_001142419.1 mRNA MORF4L2 蛋白质编码 -1.9463140034.07E-08 NM_012286.2 mRNA MORF4L2 蛋白质编码 -1.9882877482.47E-08 NM_000584.3 mRNA CXCL8 蛋白质编码 -2.6734155812.81E-11
2.3 层次聚类分析
根据差异外泌体mRNA检测结果进行层次聚类分析,分析结果如图2,其中X 轴代表进行聚类分析的差异比较组,Y轴代表样本差异外泌体mRNA。颜色代表差异倍数,越绿表示下调倍数越大,越红则表示上调倍数越大。
2.4 GO 显著性富集分析
分别对表达下调与表达上调的外泌体mRNA的靶基因做GO显著性富集分析,如图3-4,其中横坐标为差异外泌体mRNA的靶基因数,纵坐标表示GO terms。GO terms总共有3类,分别以不同颜色标注(橙色:分子功能,红色:细胞成分,绿色:生物过程)。结果显示表达下调的外泌体mRNA的靶基因与嗅觉受体活性、细胞因子活性、CXCR趋化因子受体结合、中间丝、中间丝状细胞骨架等有关;表达上调的外泌体mRNA的靶基因与蛋白质结合、蛋白质异二聚化活性、免疫系统过程的调节、胞外囊泡、细胞外细胞器、应激反应等有关。
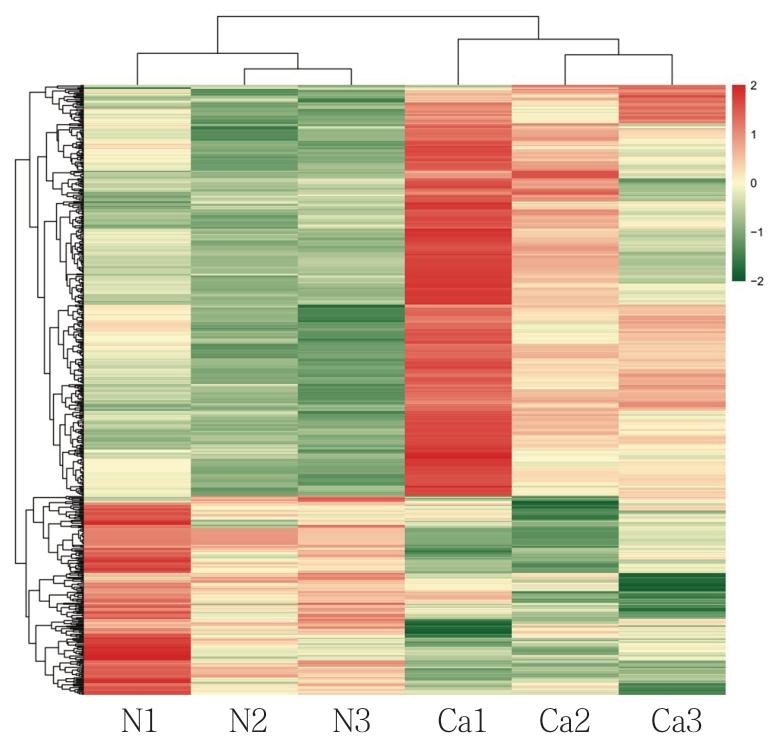
图2 差异表达外泌体mRNA 层次聚类分析
Figure2 Hierarchical clustering analysis of differentially expressed exosomal mRNAs
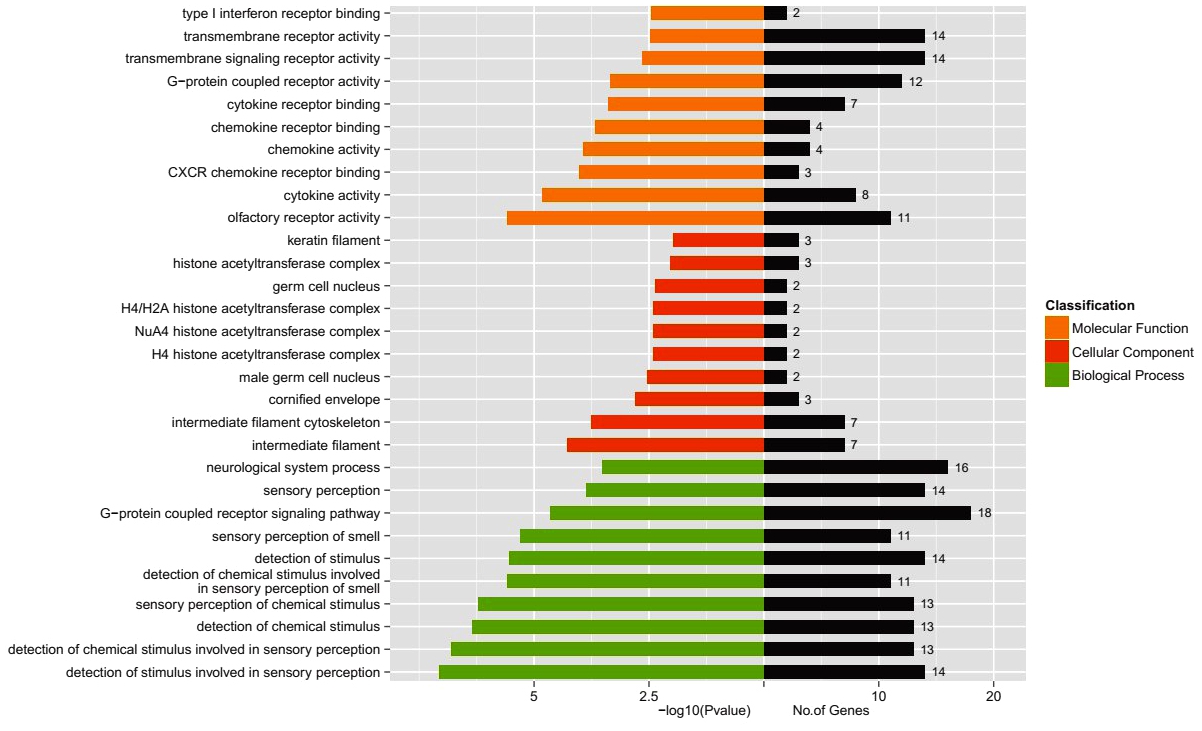
图3 表达下调mRNA 的GO 分析
Figure3 GO analysis of the down-regulated mRNAs

图4 表达上调mRNA 的GO 分析
Figure4 GO analysis of the up-regulated mRNAs
2.5 KEGG 通路显著性富集分析
分别对表达上调及下调的血清外泌体mRNA进行KEGG富集分析,结果详见图5与图6所示,横坐标表示差异外泌体mRNA的靶基因数量,纵坐标表示二级KEGG通路,图中用不同颜色表示一级通路类别。KEGG通路分析显示,在下调的外泌体mRNA中9条通路被显著富集(图5),并且在上调的外泌体mRNA中,30条通路被显著富集(图6)。 其中,在下调的外泌体mRNA中,“基础转录因子”和“细胞因子-细胞因子受体相互作用”分别是最丰富和最有意义的通路。在上调的外泌体mRNA中,“血小板激活”、“Rap 1信号通路”、“吞噬体”、“病毒致癌”、“肌动蛋白细胞骨架的调节”与“抗原处理和呈递”等是最丰富和最有意义的通路。

图5 表达下调mRNA 的KEGG 通路
Figure5 KEGG pathway analysis of the down-regulated mRNAs

图6 表达上调mRNA 的KEGG 通路
Figure6 KEGG pathway analysis of the up-regulated mRNAs
3 讨 论
外泌体作为理想的肿瘤标志物的载体,广泛分布于人体各种体液中。大量研究[18-22]已经证实其携带的蛋白质、mRNA、miRNA与lncRNA还有rRNA、tRNA、vtRNA、Y-RNA、circRNA等在细胞间进行信息交换和调节受体细胞活动,而肿瘤来源的外泌体可以调节宿主微环境,增强侵袭能力,最终导致疾病进展。有研究[23]显示,来自HCC细胞的外泌体miRNA可调节转化生长因子β活化激酶-1的表达和信号传导,促进受体细胞的生长侵袭。有学者[24]发现,HCC患者血清外泌体miR-665水平明显高于健康受试者,其升高水平与肿瘤大小、局部浸润、临床分期及生存时间密切相关,推测血清外泌体miR-665可以用于HCC诊断和判断预后。另有研究[25]结果显示,HCC患者血清外泌体miR-18a、miR-221、miR-222、miR-224水平显著高于慢性乙型肝炎(CHB)和肝硬化患者。Liu等[26]发现大鼠血清外泌体中的miRNA-10b在肝硬化即表达增加,在HCC阶段明显增加可达正常水平的10倍以上。Hoshion等[27]发现肿瘤的器官亲嗜性转移与外泌体转运整联蛋白激活受体细胞的Src基因和S100基因表达密切相关。肿瘤生长依赖于血液供应的大量营养物质,大量研究发现外泌体参与的内皮细胞迁移及血管再生对HCC的发展转移至关重要,Huang等[28]发现HCC细胞可分泌Ⅰ型跨膜蛋白VASN(vasorin)至外泌体,通过硫酸肝素蛋白聚糖(HSOGs)介导的胞吞作用转运至人脐静脉内皮细胞中促进其迁移。综上,HCC血清外泌体携带的核酸类物质是肝癌发生、发展中的重要因素,近年来对HCC外泌体miRNA研究较多[29],而mRNA研究较少,本研究通过高通量测序技术对HCC患者血清外泌体mRNA进行检测,进一步证实了外泌体内携带的肿瘤来源物质可用于HCC的早期诊断、耐药性检测、预后评估及治疗[30]。
He等[31]发现外泌体mRNA直接参与HCC的转移活动,并且还发现由外泌体激活了PI3K/ART通路,从而导致了肿瘤进展。Akiba等[32]曾发现HCC组织mRNA、蛋白质水平中的CXCL8基因表达均升高,其可增强HCC细胞和内皮细胞的运动能力,使肿瘤细胞进一步转移。CXCL8促进肿瘤血管生成的作用可能与其氨基端的ELR(Glu-Leu-Arg,谷氨酸-亮氨酸-精氨酸)序列有关[33]。BEX蛋白(brain expressed X-linked protein)的基因属于X染色体连锁基因家族[34],Braeuning等[35]发现肝癌组织中BEX1的mRNA水平比正常组织中高 400 倍,其推测BEX1 是HCC 的可靠的肿瘤标志物。趋化因子PF4(又称为血小板因子4)属于ELRCXC 趋化因子亚家族,其编码基因位于4 号染色体长臂,有研究表明趋化因子PF4 能够通过干扰血管内皮生长因子、成纤维细胞生长因子及其受体的相互作用,抑制血管新生[36]。另有研究[37]表明PF4 与其受体相互作用后,可通过c AMP/PKA通路抑制m-钙蛋白酶的活化,从而阻止细胞迁移。此外RGS18基因与G蛋白偶联受体(GPCR)通路有关,而GPCR 通路异常与肿瘤的发生和转移密切相关[38]。GPCR家族趋化因子受体在多种肿瘤的转移中具有重要作用,其异常高表达可增强肿瘤细胞的迁移能力,趋化因子在肿瘤微环境中局部释放后能通过自分泌和旁分泌途径增强癌细胞的运动和存活能 力[39],笔者推测这两种基因在HCC中同样具有重要作用。
根据上述CXCL8 及BEX、RGS18 等基因在HCC发生发展中的作用来看,本实验所筛选出来的关键基因在一定程度上是可靠的,有进一步研究的价值。我们有理由相信有可靠的外泌体mRNA会成为HCC 诊断的“金标准”,甚至未来在靶向基因治疗方面有立足之地。但本研究样本量较小,下一步拟对差异显著的mRNA在扩大样本量的血清及HCC组织中进行进一步实验验证。
[1] Chen W,Zheng R,Baade PD,et al.Cancer statistics in China,2015[J].CA Cancer J Clin,2016,66(2):115–132.doi:10.3322/caac.21338.
[2] 杜培源,宋京海,乔江春,等.肝细胞癌患者微血管侵犯影响因素分析[J].中华肝胆外科杂志,2019,25(1):26–29.doi:10.3760/cma.j.issn.1007–8118.2019.01.007.
Du PY,Song JH,Qiao JC,et al.Influencing factors analysis of microvascular invasion in patients with hepatocellular carcinoma[J].Chinese Journal of Hepatobiliary Surgery,2019,25(1):26–29.doi:10.3760/cma.j.issn.1007–8118.2019.01.007.
[3] 赵晖,陈骏,严笑鹏,等.肝细胞癌患者微血管侵犯的组织学风险分级与预后的关系[J].中华肝胆外科杂志,2019,25(6):401–405.doi:10.3760/cma.j.issn.1007–8118.2019.06.001.
Zhao H,Chen J,Yan XP,et al.Prognostic value of the risk classification of microvascular invasion in patients with hepatocellular carcinoma[J].Chinese Journal of Hepatobiliary Surgery,2019,25(6):401–405.doi:10.3760/cma.j.issn.1007–8118.2019.06.001.
[4] Torre LA,Bray F,Siegel RL,et al.Global cancer statistics,2012[J].CA Cancer J Clin,2015,65(2):87–108.doi:10.3322/caac.21262.
[5] Llovet JM,Villanueva A,Lachenmayer A,et al.Advances in targeted therapies for hepatocellular carcinoma in the genomic era[J].Nat Rev Clin Oncol,2015,12(8):436.doi:10.1038/nrclinonc.2015.121.
[6] Bruix J,Reig M,Sherman M.Evidence-Based Diagnosis,Staging,and Treatment of Patients With Hepatocellular Carcinoma[J].Gastroenterology,2016,150(4):835–853.doi:10.1053/j.gastro.2015.12.041.
[7] Kulik LM,Chokechanachaisakul A.Evaluation and Management of Hepatocellular Carcinoma[J].Clin Liver Dis,2015,19(1):23–43.doi:10.1016/j.cld.2014.09.002.
[8] Das T,Prodhan C,Patsa S,et al.Identification of over expressed proteins in oral submucous fibrosis by proteomic analysis[J].J Cell Biochem,2018,119(6):4361–4371.doi:10.1002/jcb.26423.
[9] 孙梦雨,邱洁萍,吴之涵,等.胃癌枢纽基因的筛选和预后分析[J].中国普通外科杂志,2019,28(4):433–440.doi:10.7659/j.issn.1005–6947.2019.04.008.
Sun MY,Qiu JP,Wu ZH,et al.Identification and prognostic analysis of hub genes in gastric cancer[J].Chinese Journal of General Surgery,2019,28(4):433–440.doi:10.7659/j.issn.1005–6947.2019.04.008.
[10] 白文萱,高健,钱程,等.肝癌相关差异表达基因的生物信息学分析[J].中华肝脏病杂志,2017,25(6):435–439.doi:10.3760/cma.j.issn.1007–3418.2017.06.009.
Bai WX,Gao J,Qian C,et al.A bioinformatics analysis of differentially expressed genes associated with liver cancer[J].Chinese Journal of Hepatology,2017,25(6):435–439.doi:10.3760/cma.j.issn.1007–3418.2017.06.009.
[11] Zhang C,Peng L,Zhang Y,et al.The identification of key genes and pathways in hepatocellular carcinoma by bioinformatics analysis of high-throughput data [J].Med Oncol,2017,34(6):101.doi:10.1007/s12032–017–0963–9.
[12] 王兴春,杨致荣,王敏,等.高通量测序技术及其应用[J].中国生物工程杂志,2012,32(1):109–114.
Wang XC,Yang ZR,Wang M,et al.High-throughput Sequencing Technology and Its Application[J].China Biotechnology,2012,32(1):109–114.
[13] Altanerova U,Jakubechova J,Repiska V,et al.Exosomes of human mesenchymal stem/stromal/medicinal signaling cells[J].Neoplasma,2017,64(6):809–815.doi:10.4149/neo_2017_601.
[14] Sugimachi K,Matsumura T,Hirata H,et al.Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation[J].Br J Cancer,2015,112(3):532–538.doi:10.1038/bjc.2014.621.
[15] Pan JH,Zhou H,Zhao XX,et al.Role of exosomes and exosomal microRNAs in hepatocellular carcinoma:Potential in diagnosis and antitumour treatments (Review)[J].Int J Mol Med,2018,41(4):1809–1816.doi:10.3892/ijmm.2018.3383.
[16] Wang Y,Zhang C,Zhang P,et al.Serum exosomal microRNAs combined with alpha-fetoprotein as diagnostic markers of hepatocellular carcinoma [J].Cancer Med,2018,7(5):1670–1679.doi:10.1002/cam4.1390..
[17] Tang J,Li Y,Liu K.Exosomal miR-9–3p suppresses HBGF-5 expression and is a functional biomarker in hepatocellular carcinoma[J].Minerva Med,2018,109(1):15–23.doi:10.23736/S0026–4806.17.05167–9.
[18] Lu D,Xu AD.Mini Review:Circular RNAs as Potential Clinical Biomarkers for Disorders in the Central Nervous System[J].Front Genet,2016,7:53.doi:10.3389/fgene.2016.00053.
[19] 刘鹏,刘合利.结直肠癌液体活检的临床应用进展[J].中国普通外科杂志,2019,28(9):1143–1149.doi:10.7659/j.issn.1005–6947.2019.09.017.
Liu P,Liu HL.Advances in clinical applications of liquid biopsies for colorectal cancer[J].Chinese Journal of General Surgery,2019,28(9):1143–1149.doi:10.7659/j.issn.1005–6947.2019.09.017.
[20] Simons M,Raposo G.Exosomes--vesicular carriers for intercellular communication[J].Curr Opin Cell Biol,2009,21(4):575–581.doi:10.1016/j.ceb.2009.03.007.
[21] Mathivanan S,Ji H,Simpson RJ.Exosomes:extracellular organelles important in intercellular communication[J].J Proteomics,2010,73(10):1907–1920.doi:10.1016/j.jprot.2010.06.006.
[22] Kahlert C,Kalluri R.Exosomes in tumor microenvironment influence cancer progression and metastasis[J].J Mol Med (Berl),2013,91(4):431–437.doi:10.1007/s00109–013–1020–6.
[23] Kogure T,Lin WL,Yan IK,et al.Intercellular nanovesicle-mediated microRNA transfer:a mechanism of environmental modulation of hepatocellular cancer cell growth[J].Hepatology,2011,54(4):1237–1248.doi:10.1002/hep.24504.
[24] Qu Z,Wu J,Wu J,et al.Exosomal miR-665 as a novel minimally invasive biomarker for hepatocellular carcinoma diagnosis and prognosis[J].Oncotarget,2017,8(46):80666–80678.doi:10.18632/oncotarget.20881.
[25] Sohn W,Kim J,Kang SH,et al.Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma[J].Exp Mol Med,2015,47:e184.doi:10.1038/emm.2015.68.
[26] Liu WH,Ren LN,Wang X,et al.ombination of exosomes and circulating microRNAs may serve as a promising tumor marker complementary to alpha-fetoprotein for early-stage hepatocellular carcinoma diagnosis in rats [J].J Cancer Res Clin Oncol,2015,141(10):1767–1778.doi:10.1007/s00432–015–1943–0.
[27] Hoshino A,Costa-Silva B,Shen TL,et al.Tumour exosome integrins determine organotropic metastasis[J].Nature,2015,527(7578):329–335.doi:10.1038/nature15756.
[28] Huang A,Dong J,Li S,et al.Exosomal transfer of vasorin expressed in hepatocellular carcinoma cells promotes migration of human umbilical vein endothelial cells[J].Int J Biol Sci,2015,11(8):961–969.doi:10.7150/ijbs.11943.
[29] 高斌,熊莹晖,黄泽炳,等.乙肝相关性肝细胞癌患者血清外泌体miR-1290水平的变化及其诊断价值[J].中国普通外科杂志,2019,28(1):31–38.doi:10.7659/j.issn.1005–6947.2019.01.005.
Gao B,Xiong YH,Huang ZB,et al.Change in serum level of exosomal miR-1290 in patients with hepatitis B virus related hepatocellular carcinoma and its diagnostic value[J].Chinese Journal of General Surgery,2019,28(1):31–38.doi:10.7659/j.issn.1005–6947.2019.01.005.
[30] Moris D,Beal EW,Chakedis J,et al.Role of exosomes in treatment of hepatocellular carcinoma[J].Surg Oncol,2017,26(3):219–228.doi:10.1016/j.suronc.2017.04.005.
[31] He M,Qin H,Poon TC,et al.Hepatocellular carcinoma-derived exosomes promote motility of immortalized hepatocyte through transfer of oncogenic proteins and RNAs[J].Carcinogenesis,2015,36(9):1008–1018.doi:10.1093/carcin/bgv081.
[32] Akiba J1,Yano H,Ogasawara S,et al.Expression and function of interleukin-8 in human hepatocellular carcinoma[J].Int J Oncol,2001,18(2):257–264.doi:10.3892/ijo.18.2.257.
[33] Bizzarri C,Beccari AR,Bertini R,et al.ELR+ CXC chemokines and their receptors (CXC chemokine receptor 1 and CXC chemokine receptor 2) as new therapeutic targets[J].Pharmacol Ther,2006,112(1):139–149.doi:10.1016/j.pharmthera.2006.04.002.
[34] 王永,于如同,孟庆明.Bex2抑制神经胶质瘤细胞凋亡及其与 JNK/MAPK 通路关系的研究[J].中国药物与临床,2014,14(10):1309–1312.doi:10.11655/zgywylc2014.10.001.
Wang Y,Yu RT,Meng QM.The inhibitory effect of Bex2 on apoptosis of glioma cells and the association with JNK/MAPK pathway[J].Chinese Remedies & Clinics,2014,14(10):1309–1312.doi:10.11655/zgywylc2014.10.001.
[35] Braeuning A,Jaworski M,Schwarz M,et al.Rex3 (reduced in expression 3) as a new tumor marker in mouse hepatocarcinogenesis[J].Toxicology,2006,227(1–2):127–135.doi:10.1016/j.tox.2006.07.024.
[36] Struyf S,Burdick MD,Peeters E,et al.Platelet factor-4 variant chemokine CXCL4L1 inhibits melanoma and lung carcinoma growth and metastasis by preventing angiogenesis[J].Cancer Res,2007,67(12):5940–5948.doi:10.1158/0008–5472.CAN-06–4682.
[37] Wu Q,Dhir R,Wells A.Altered CXCR3 isoform expression regulates prostate cancer cell migration and invasion[J].Mol Cancer,2012,11:3.doi:10.1186/1476–4598–11–3.
[38] Li DQ,Pakala SB,Nair SS,et al.Metastasis-associated protein 1/nucleosome remodeling and histone deacetylase complex in cancer[J].Cancer Res,2012,72(2):387–394.doi:10.1158/0008–5472.CAN-11–2345.
[39] Sen N,Gui B,Kumar R.Role of MTA1 in cancer progression and metastasis[J].Cancer Metastasis Rev,2014,33(4):879–889.doi:10.1007/s10555–014–9515–3.
