胰腺癌是一种高病死率疾病,其恶性程度高,早期诊断困难,手术切除率低,对放化疗不敏感,容易复发和远处转移[1]。因此,探索胰腺癌发生发展相关机制,寻找诊治新靶标,具有重要意义。
尿苷二磷酸-葡萄糖6-脱氢酶(UDP-glucose 6-dehydrogenase,UGDH)编码的蛋白将UDP-葡萄糖转化为UDP-葡萄糖醛酸。UDP-葡萄糖醛酸是糖胺聚糖(GAG)包括透明质酸(HA)、硫酸软骨素(CS)和硫酸肝素(HS)葡萄糖醛酸化和合成的底物。这些糖基化化合物是细胞外基质的常见成分,可能在信号转导、细胞迁移、癌症生长中发挥作用[2]。目前已经有越来越多证据表明UGDH参与肺腺癌、胶质母细胞瘤、结直肠癌、前列腺癌、乳腺癌等多种癌症的发生和发展。Wang等[3]发现了UDP-葡萄糖在肺癌转移中的肿瘤抑制作用,UGDH通过增加SNAI1mRNA的稳定性促进肿瘤转移。Oyinlade等[4]发现类Krüppel因子4(KLF4)通过与甲基化DNA结合,使基因表达活化来促进胶质母细胞瘤细胞的迁移,UDGH是KLF4-mCpG结合活性的下游靶点之一,开发UGDH拮抗剂以减少细胞外基质成分的合成是治疗胶质母细胞瘤的有效策略。Wang等[5]发现UGDH的下调影响HCT-8结直肠癌细胞中GAG的合成和运动,该酶可能是结直肠癌治疗干预的潜在靶点。Huang等[6]的研究也表明UGDH是一种新型的前列腺癌特异性生物标志物。Arnold等[7]研究发现UGDH调节HA的产生并促进乳腺癌的进展,上皮-间质转化(EMT)可以促进代谢重编程,从而通过氨基糖和核苷酸糖通路增加葡萄糖向HA的通量。Theocharis等[8]在一项研究中发现,相比于人的正常胰腺组织,胰腺癌组织的总GAG含量增加4倍,HA增加12倍,CS增加22倍;HA是肿瘤浸润性生长的关键分子之一,高水平的HA与某些肿瘤较差分化和低存活率相关;CS在各种间质和上皮肿瘤中显著增高,其积累可能与肿瘤生长和进展相关。HA与其细胞表面受体CD44和RHAMM的相互作用,促进肿瘤细胞生长、存活和迁移,从而增加转移扩散[9]。因此,UGDH可能通过参与GAG在肿瘤组织中的生物合成来影响肿瘤的侵袭能力和耐药性[10]。
目前尚无胰腺癌中关于UGDH基因的研究。本研究通过生物信息学方法分析UGDH与胰腺癌的生存预后,及其与相关基因的共表达分析,并利用GSEA分析及单基因PPI预测等方法,以期揭示UGDH在胰腺癌发生发展中的作用。
1 材料与方法
1.1 UGDH 基因在正常胰腺组织和胰腺癌组织中的表达差异
基于Oncomine[11]数据库,分析UGDH基因在不同数据集中,在胰腺癌样本和正常样本中的差异表达情况。
1.2 UGDH 表达与胰腺癌患者预后之间的关系
首先在UALCAN[12]数据库中直接查看UGDH基因的高低表达与胰腺癌预后之间的关系,然后使用R语言和从UCSC Xena下载得到的TCGA中胰腺癌基因表达谱和生存数据进行生存分析。首先从生存时间的样本中,将正常样本去除;然后将生存状态为0(存活)且随访时间<30 d的样本视为随访失败去除。经以上处理后得样本,提取其基因表达量,使用R语言的Survminer[13]包计算最佳截断值(optimal cut-point),然后以该值作为阈值,高于该值作为高表达,低于该值作为低表达,与生存时间和生存状态一起进行生存分析。
1.3 UGDH 基因与其他基因的共表达分析及在胰腺癌中可能参与、调控的生物学功能分析
基于UALCAN 数据库,分析TCGA 中胰腺癌UGDH 基因与其他基因的表达相关性,使用Pearson系数法,获得其显著正相关与负相关的基因,分别进行KEGG通路和GO(BP、CC、MF)的富集分析,富集分析采用R语言ClusterProfiler[14]包进行。
1.4 基因集富集分析(gene set enrichment analysis,GSEA)
基于HGNC数据库提供的人类所有基因的注释,提取胰腺癌中所有mRNA的表达谱,然后基于GSEA分析软件,进行UGDH基因在胰腺癌中的基因基富集分析,获得UGDH基因的正向和负向调控通路,以结果中P<0.05的结果作为显著富集结果,表达谱已经过Log2(count+1)的标准化转化,以消除量纲差异。
1.5 UGDH 与其他基因的作用关系预测
基于HGNC 数据库提供的人类所有基因的注释,提取胰腺癌中所有m RNA 的表达谱基于GeneMANIA[15]对UGDH基因进行单基因蛋白互作(PPI)预测分析,进一步对网络中出现的基因使用Metascape[16]进行功能富集分析;使用Cytoscape的MCODE插件对网络进行关键子网络分析。
2 结 果
2.1 UGDH 基因在正常胰腺组织和胰腺癌组织中的表达差异
通过Oncomine数据库的4个胰腺癌数据集,发现胰腺癌样本和正常样本中,UGDH基因的表达出现显著差异,且在胰腺癌中UGDH呈高表达(均P<0.05)(图1)。
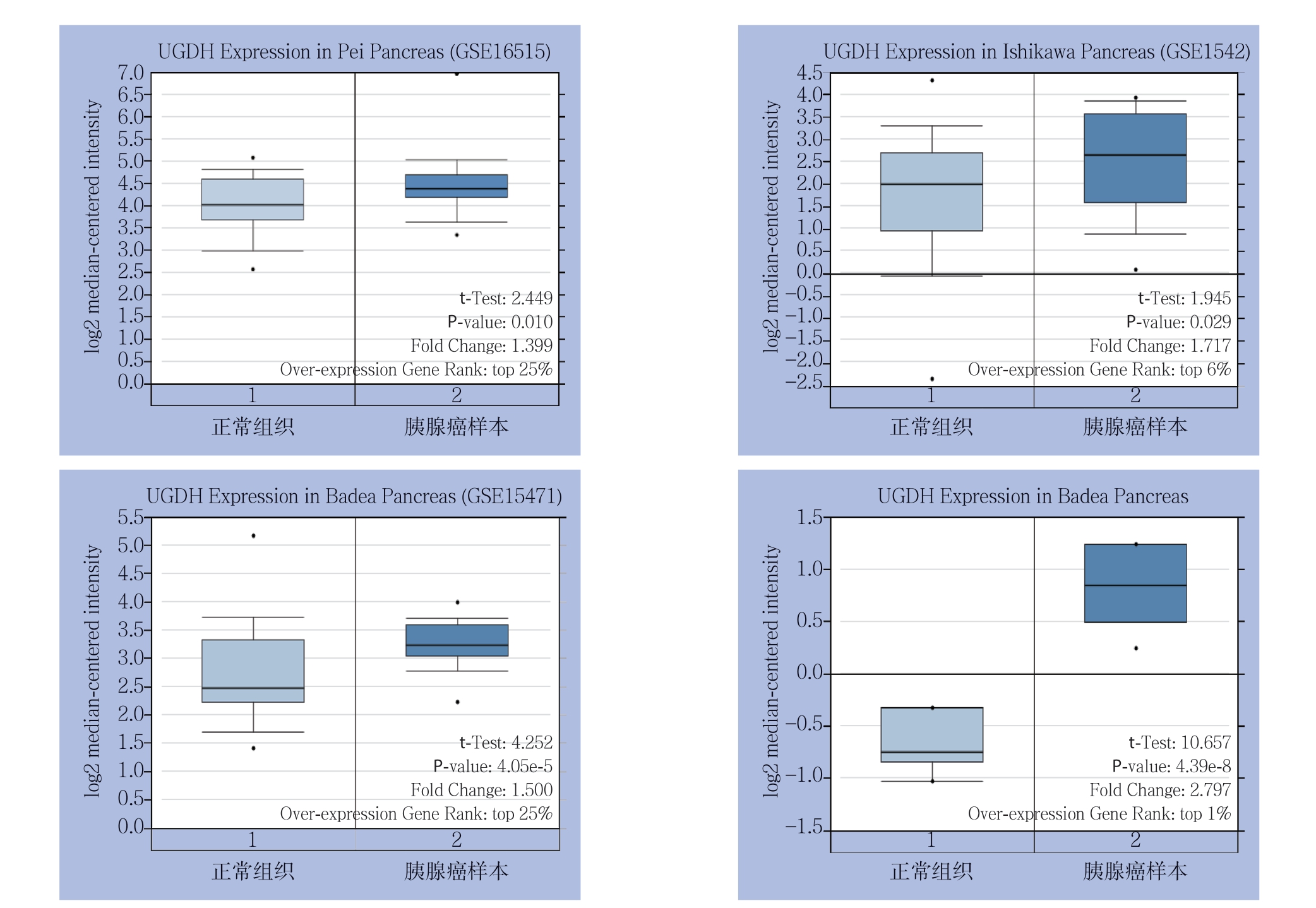
图1 不同数据集中UGDH 在胰腺癌样本和正常组织中的表达
Figure1 UGDH expressions in pancreatic cancer samples and normal tissues in different data sets
2.2 UGDH 表达与胰腺癌患者预后之间的关系
生存分析结果发现,UGDH基因的表达水平对胰腺癌患者的总生存率有明显影响。与UGDH高表达组相比,UGDH低表达组的胰腺癌患者其总生存率明显升高,生存期更长(均P<0.05)(图2)。
2.3 UGDH 基因与其他基因的共表达及在胰腺癌中可能参与、调控的生物学功能分析
基于UALCAN 数据库,分析TCGA 中胰腺癌UGDH 基因与其他基因的表达相关性,使用Pearson系数法,获得其显著正相关与负相关的基因,部分结果如表1所示。通过GO富集分析了解差异基因UGDH富集于内膜系统、RNA定位、蛋白酶体蛋白分解代谢过程、核质转运、核酸运输等与癌症相关生物学过程;KEGG通路富集分析可见差异基因显著富集于内质网蛋白加工、自噬、泛素介导的蛋白水解、蛋白质输出、剪接体、RNA转运、mRNA监测通路、病毒致癌作用、长期抑制作用、结直肠癌、N-聚糖的生物合成、胰腺癌通路、鞘脂信号通路、癌症中的蛋白聚糖、肾细胞癌、人类巨细胞病毒感染、乙肝等与癌症相关通路(均P<0.05)。相关研究表明,核质转运、核酸运输、高尔基体和外周囊泡及侵袭性GTPase信号传导、自噬作用、mRNA监测通路(如无义介导的mRNA衰变)、N-聚糖的生物合成、胰腺癌通路、鞘脂信号通路在胰腺癌的发生发展中均有显著意义[17-23]。结果表明,与UGDH显著共表达的基因,富集到多条癌症相关生物学功能及信号通路,其中包含胰腺癌通路,这说明UGDH基因在胰腺癌中发挥重要作用(图3)。
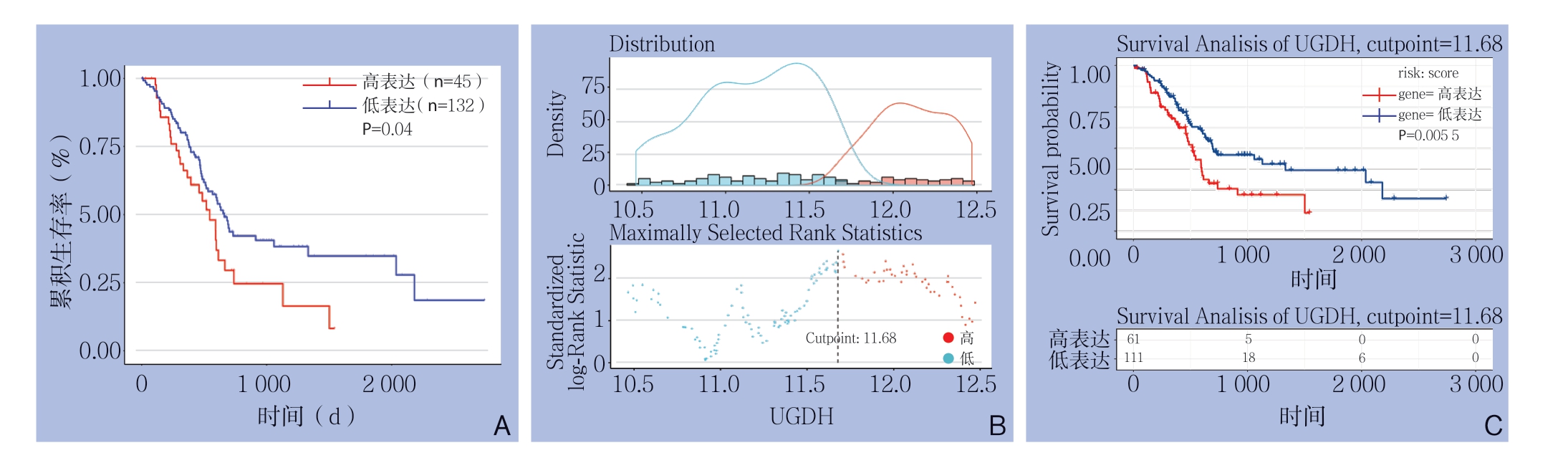
图2 胰腺癌中UGDH 表达与预后的关系 A:UALCAN 数据库中胰腺癌UGDH 表达对预后作用的生存分析;B:Survminer包计算得到的最佳截断值;C:基于最佳截断值进行的TCGA 中UGDH 与胰腺癌关系的生存分析
Figure 2 Relationship between UGDH expression in pancreatic cancer and the prognosis A: Survival analysis of the effect of UGDH expression on prognosis in UALCAN database; B: The optimal cut point obtained by using Survminer package; C: Survival analysis of the relationship between UGDH and pancreatic cancer in TCGA database
表1 TCGA 数据库胰腺癌UGDH 基因与其他基因的共表达关系
Table 1 Coexpression relationship between UGDH gene and other genes in pancreatic cancer in TCGA database
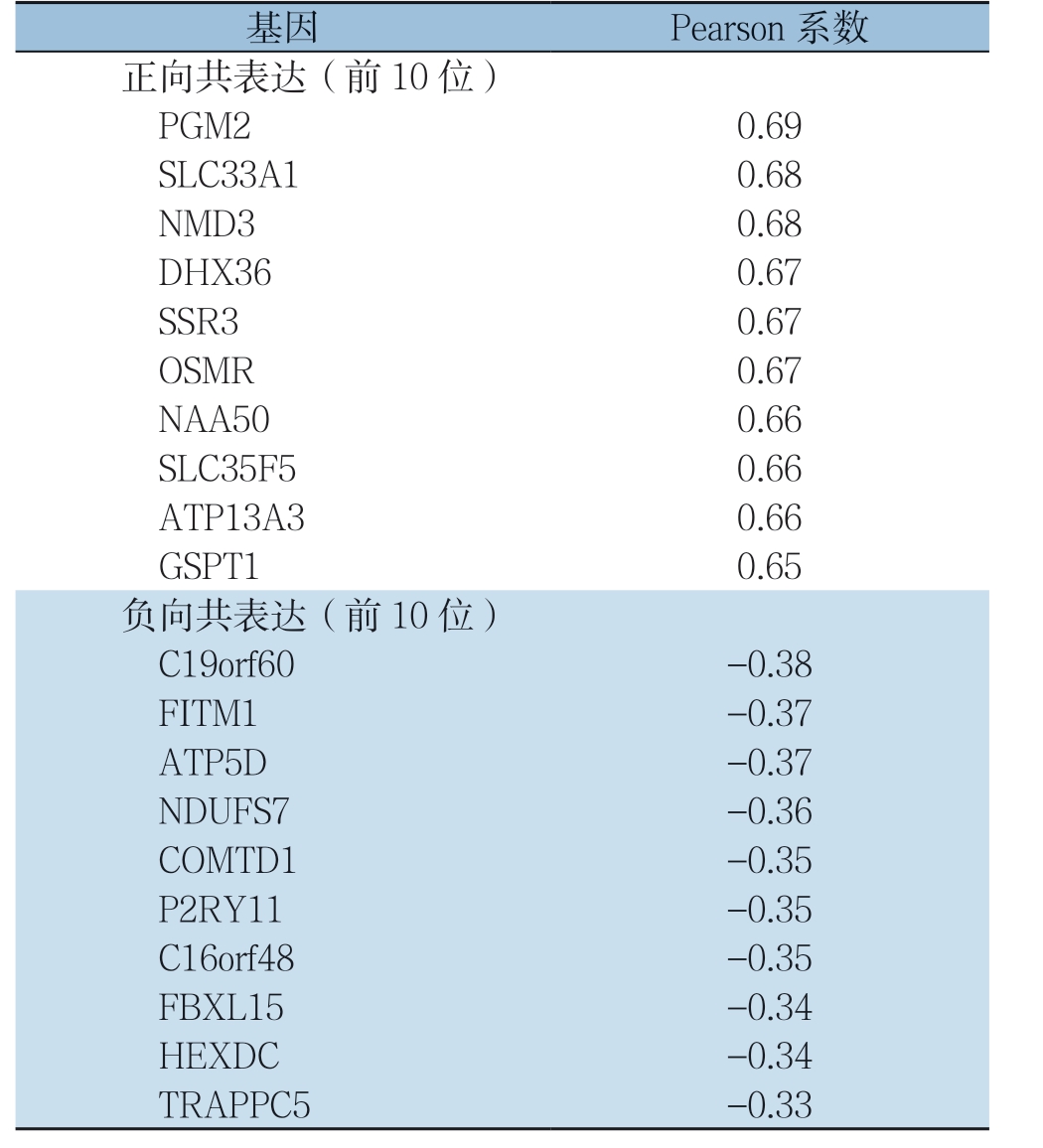
基因 Pearson 系数正向共表达(前10 位)PGM2 0.69 SLC33A1 0.68 NMD3 0.68 DHX36 0.67 SSR3 0.67 OSMR 0.67 NAA50 0.66 SLC35F5 0.66 ATP13A3 0.66 GSPT1 0.65负向共表达(前10 位)C19orf60 -0.38 FITM1 -0.37 ATP5D -0.37 NDUFS7 -0.36 COMTD1 -0.35 P2RY11 -0.35 C16orf48 -0.35 FBXL15 -0.34 HEXDC -0.34 TRAPPC5 -0.33
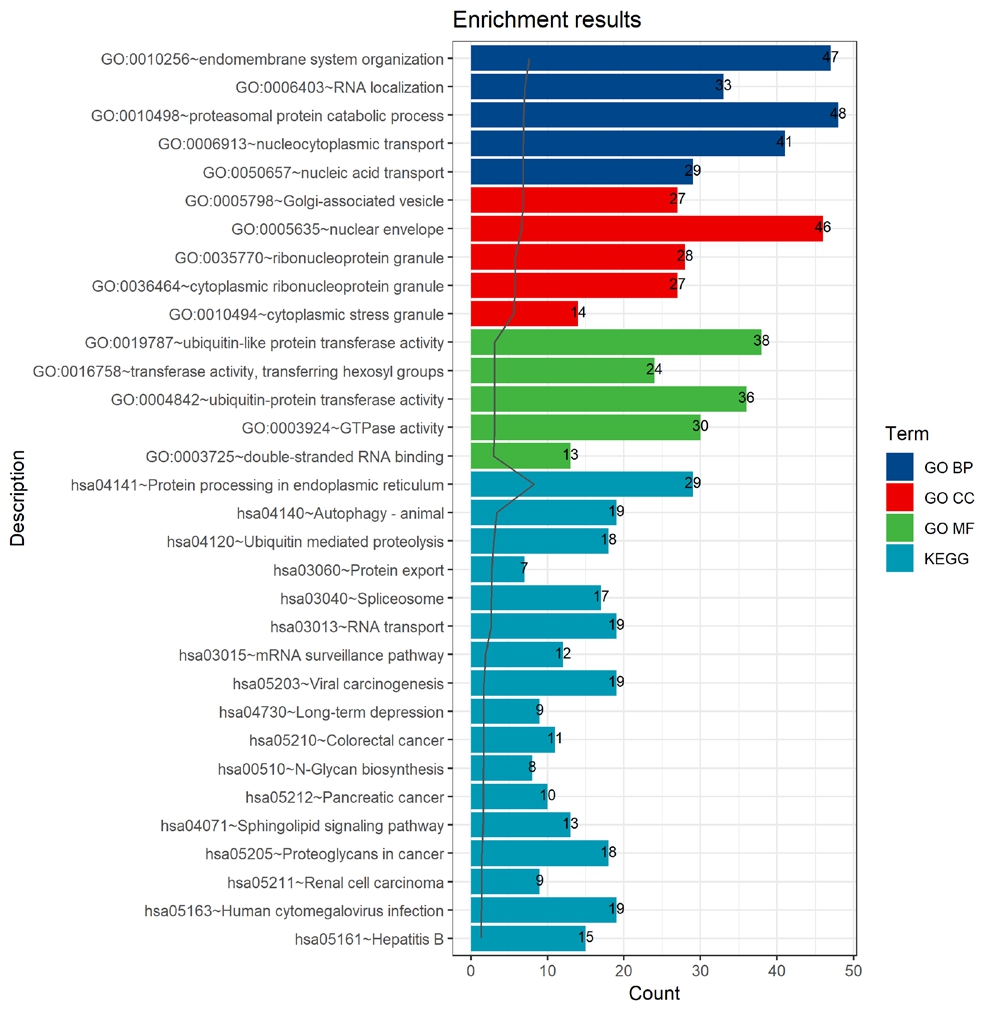
图3 胰腺癌中与UGDH 基因显著共表达的基因的功能和通路富集条形图
Figure 3 Function and pathway enrichment bar graph of genes significantly co-expressed with UGDH gene in pancreatic cancer
2.4 GSEA 分析
GSEA分析是一种针对全基因组表达谱芯片数据的分析方法,用来评估一个预先定义的基因集的基因在与表型相关度排序的基因表中的分布趋势,从而判断其对表型的贡献。如表2与图4所示,U G D H 在胰腺癌中的G S E A 分析富集到以下与癌症相关通路:泛素介导的蛋白水解作用、氨基糖和核苷酸糖代谢、氨酰生物合成、卵母细胞减数分裂、肾细胞癌、基础转录因子、鞘脂类代谢、ErbB信号通路、黏附连接、慢性粒细胞白血病、RIG-I样受体信号传导通路、促性腺激素释放激素信号通路、胰岛素信号通路、子宫内膜癌、神经胶质瘤、神经营养蛋白信号通路、内吞作用、MAPK信号通路、胰腺癌等(均P<0.05)。有多项研究表明促性腺激素释放激素、神经营养蛋白及MAPK信号通路在胰腺癌发生发展中起重要作用[24-26]。结果显示,UGDH基因GSEA与共表达分析均富集到多条癌症相关通路,并且均包含胰腺癌通路,进一步确定了UGDH在胰腺癌中的作用(表2)(图4)。
表2 GSEA 分析结果中明显富集的通路
Table 2 Significantly enriched pathways from GSEA analysis results
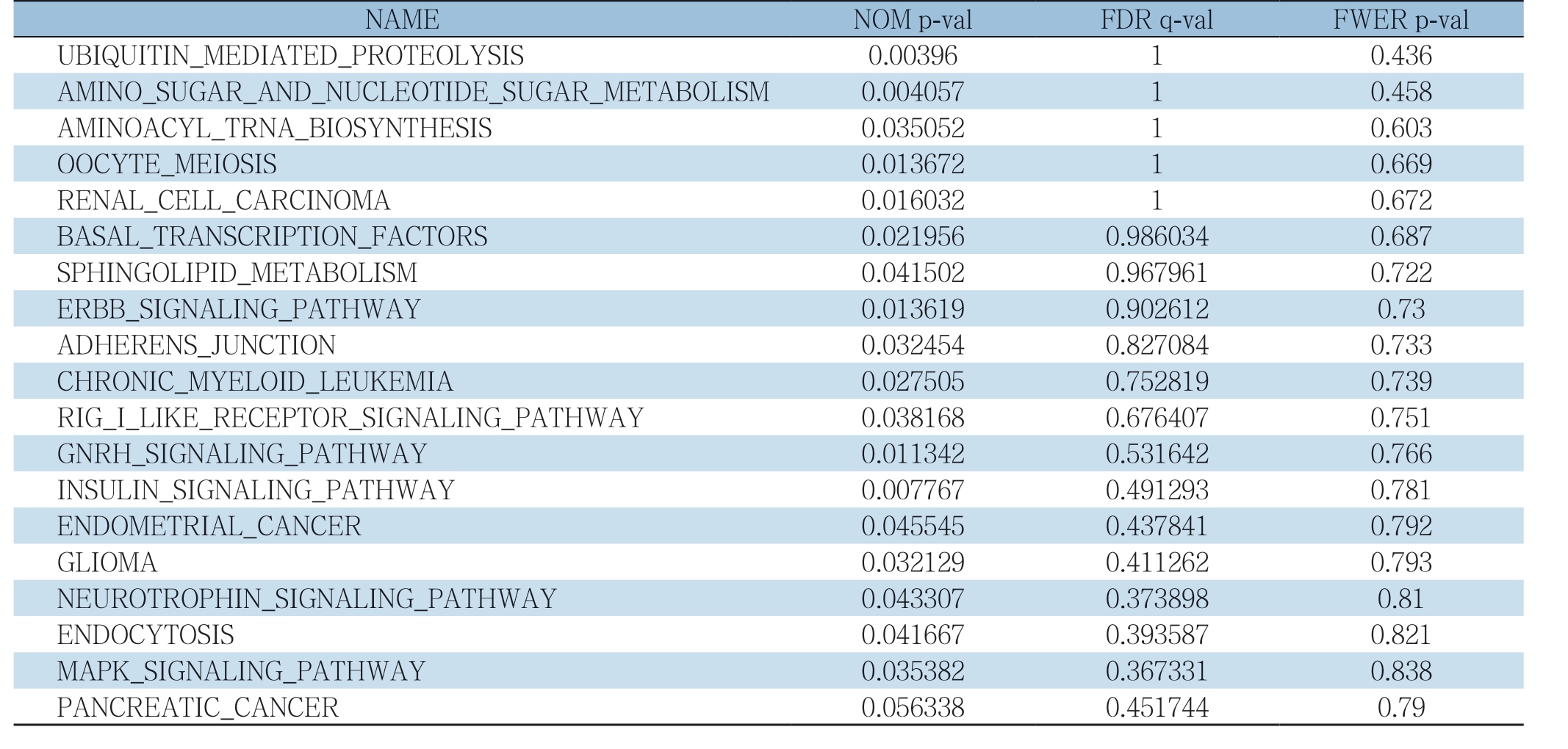
NAME NOM p-val FDR q-val FWER p-val UBIQUITIN_MEDIATED_PROTEOLYSIS 0.00396 1 0.436 AMINO_SUGAR_AND_NUCLEOTIDE_SUGAR_METABOLISM 0.004057 1 0.458 AMINOACYL_TRNA_BIOSYNTHESIS 0.035052 1 0.603 OOCYTE_MEIOSIS 0.013672 1 0.669 RENAL_CELL_CARCINOMA 0.016032 1 0.672 BASAL_TRANSCRIPTION_FACTORS 0.021956 0.986034 0.687 SPHINGOLIPID_METABOLISM 0.041502 0.967961 0.722 ERBB_SIGNALING_PATHWAY 0.013619 0.902612 0.73 ADHERENS_JUNCTION 0.032454 0.827084 0.733 CHRONIC_MYELOID_LEUKEMIA 0.027505 0.752819 0.739 RIG_I_LIKE_RECEPTOR_SIGNALING_PATHWAY 0.038168 0.676407 0.751 GNRH_SIGNALING_PATHWAY 0.011342 0.531642 0.766 INSULIN_SIGNALING_PATHWAY 0.007767 0.491293 0.781 ENDOMETRIAL_CANCER 0.045545 0.437841 0.792 GLIOMA 0.032129 0.411262 0.793 NEUROTROPHIN_SIGNALING_PATHWAY 0.043307 0.373898 0.81 ENDOCYTOSIS 0.041667 0.393587 0.821 MAPK_SIGNALING_PATHWAY 0.035382 0.367331 0.838 PANCREATIC_CANCER 0.056338 0.451744 0.79
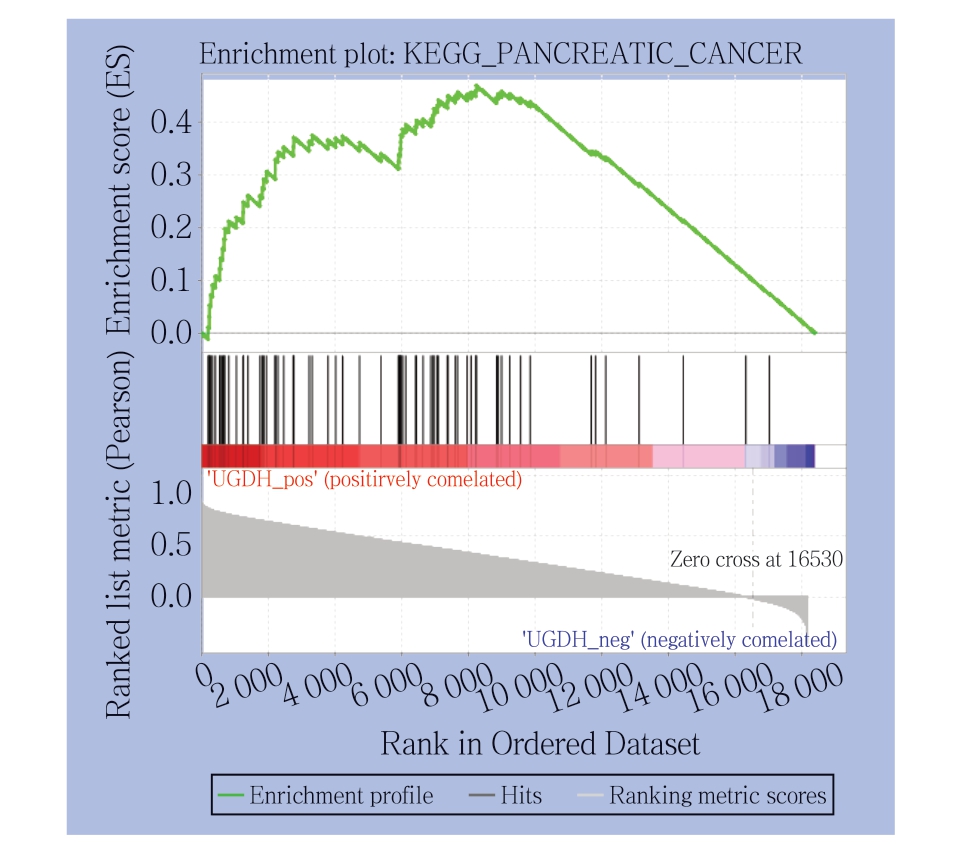
图4 GSEA 富集分析得到的胰腺癌通路
Figure 4 The pancreatic cancer pathway enriched by GSEA analysis
2.5 UGDH 与其他基因的作用关系预测
单基因PPI预测分析,使用Cytoscape进行关键子网络分析,其中HGS、Ube2v1、MAT2A、SUMO1 在互作网络中有重要作用,其表达与U G D H 基因表达呈明显正相关。Ube2v1 在表观遗传学上抑制结直肠癌中的自噬基因表达,促进转移[27]。MAT2A在肝癌[28]、结直肠癌[29]、耐他莫昔芬的乳腺癌[30]细胞中的基本表达水平上调。lncRNAs SUMO1P3可能通过EMT信号通路促进胰腺癌的细胞增殖,迁移和侵袭[31]。通过Metascape对网络中的基因进行的功能和通路富集分析,发现这些基因富集于蛋白质在膜中的定位、碳水化合物衍生物生物合成过程、蛋白质分解代谢过程的调控、肽基赖氨酸修饰等生物学过程及第二阶段-化合物的结合通路(图5-6)。细胞内蛋白的调节和降解主要是由泛素-蛋白酶体系统结合分子伴侣、自噬和溶酶体蛋白水解进行的[32]。一项研究表明,赖氨酰氧化同源物2(LOXL2)在AsPC-1和BxPC-3细胞中的过表达增强了EMT样过程,并增加了迁移和侵袭活性,且80例胰腺癌患者样本的免疫组织化学分析显示,81.2%患者胰腺癌组织中LOXL2高表达[33]。因此,UGDH有可能通过影响上述关键因子调控胰腺癌的发生发展。
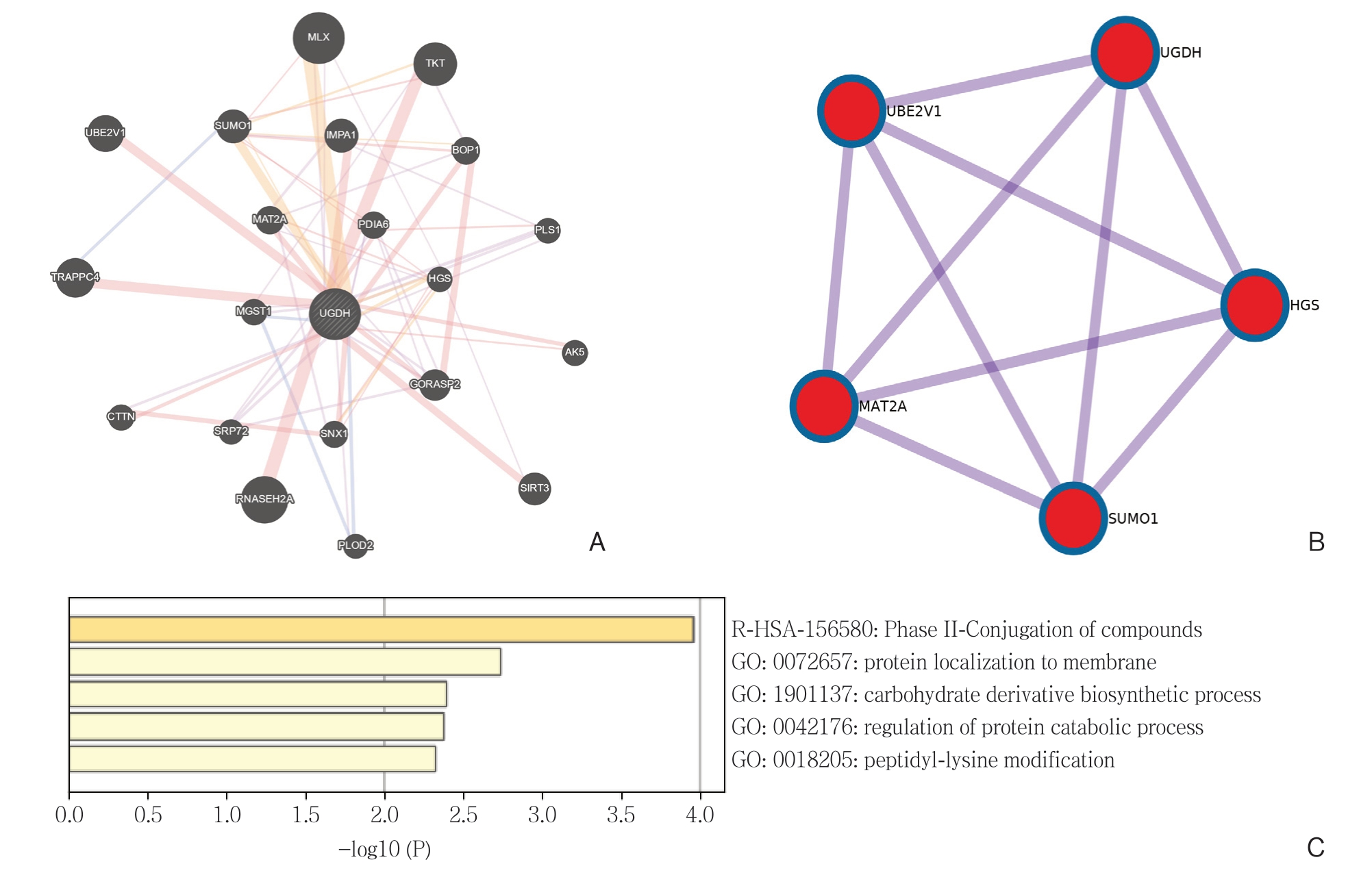
图5 UGDH 与其他基因相互作用关系分析 A:GeneMANIA 对UGDH 与其他基因的PPI 分析结果;B:Cytoscape 的MCODE 插件对网络进行关键子网络分析得到的关键子网络;C:Metascape 对网络中的基因进行的功能和通路富集分析结果
Figure 5 Analysis of the interactions of UGDH with other genes A: Results of PPI analysis of UGDH and other genes constructed by GeneMANIA; B: The key subnetworks identified in the PPI network by using Cytoscape MCODE plugin; C: Function and pathway enrichment analysis of genes in the network by using Metascape
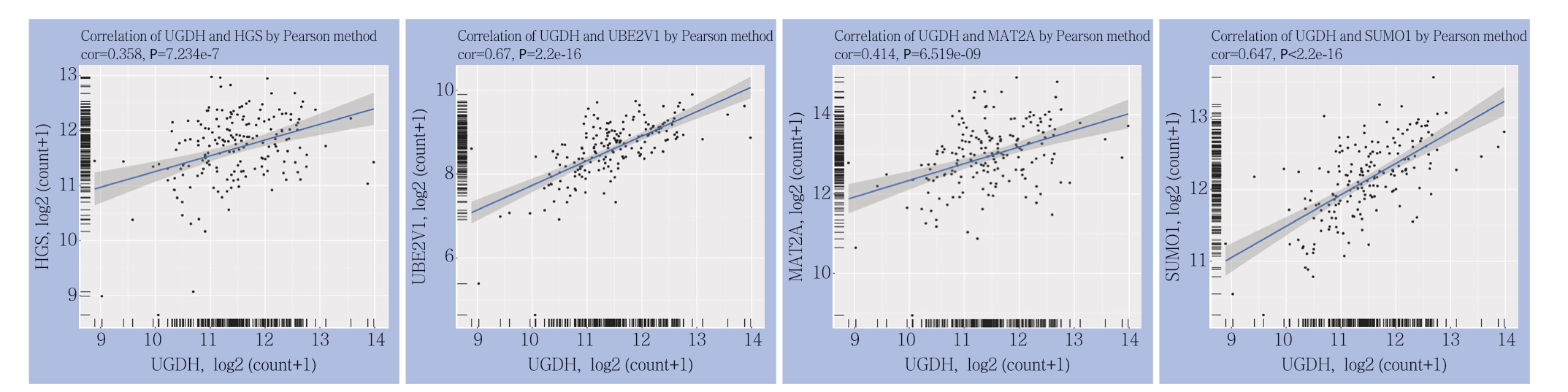
图6 PPI 分析关键子网络与UGDH 基因共表达关系
Figure 6 Co-expression relationship between the key subnetworks of PPI analysis and UGDH gene
3 讨 论
UGDH基因在肿瘤发生发展过程中起重要作用,通过UGDH靶向透明质酸的生成可以抑制多种癌症病理过程,尤其是通过限制UDP-葡萄糖醛酸的生成抑制肿瘤生长[7]。UGDH基因高表达与化疗耐药性有关[34]。需要认识到,现有的实验证据远远不足以揭开UGDH的谜底,诸如胰腺癌等癌症与UGDH之间的关系尚未阐明。siUGDH是阐明人类UGDH功能的有效工具[5],有必要进一步分析UGDH参与的Akt相关通路中的上下游分子[35]及对核UGDH进行功能研究[3]。UGDH抑制剂的开发仍处于起步阶段。未来针对UDGH的小分子抑制剂的研究将检验UGDH是否可以单独作为治疗靶点[4]。除了结合功效和抑制效力外,UGDH抑制剂的未来设计方向应集中于选择性(例如UGDH-I与UGDH-II)和细胞靶向方面。
本文利用已有的生物信息学资源和工具进行数据处理和分析,结果显示,胰腺癌样本和正常样本中,UGDH基因的表达出现显著差异,在胰腺癌组织中UGDH显著高表达;且与UGDH高表达组相比,UGDH低表达组的胰腺癌患者其总生存率显著升高,生存期更长;UGDH基因GSEA与共表达分析均富集到多条胰腺癌相关通路,进一步确定了UGDH在胰腺癌中的作用;单基因PPI预测分析显示 SUMO1等在互作网络中有重要作用,并且lncRNAs SUMO1P3可能通过EMT信号通路促进胰腺癌的细胞增殖,迁移和侵袭[31]。有研究[26]表明,钙网蛋白通过整合素/EGFR-ERK/MAPK信号通路促进胰腺癌细胞中表皮生长因子诱导的EMT。GSEA分析富集到的氨基糖和核苷酸糖代谢通路在UGDH与乳腺癌的作用机制中起重要作用,富集到的MAPK信号通路在胰腺癌中起重要作用;综上,SUMO1及EMT信号通路、MAPK信号通路、氨基糖和核苷酸糖代谢通路与UGDH之间的作用机制值得我们进一步研究。
生物信息学分析为UGDH在胰腺癌发生发展中的作用提供了全新的视角,不仅可以获得关于基因功能研究的重要线索,更为下一步实验策略提供指导。当然这种完全基于计算机的基因虚拟数字化表达研究方法也存在一定局限性。首先,它是依托于数据库,其数据的准确性决定了结果的准确性。其次,本研究缺少基础实验的验证,后续研究将继续从mRNA及蛋白质层面完善。
[1] Kamisawa T, Wood LD, Itoi T, et al.Pancreatic cancer[J].Lancet,2016, 388(10039):73-85.doi: 10.1016/S0140-6736(16)00141-0.
[2] Basappa, Rangappa KS, Sugahara K.Roles of glycosaminoglycans and glycanmimetics in tumor progression and metastasis[J].Glycoconj J, 2014, 31(6/7):461-467.doi: 10.1007/s10719-014-9551-9.
[3] Wang X, Liu R, Zhu W, et al.UDP-glucose accelerates SNAI1 mRNA decay and impairs lung cancer metastasis[J].Nature, 2019,571(7763):127-131.doi: 10.1038/s41586-019-1340-y.
[4] Oyinlade O, Wei S, Lal B, et al.Targeting UDP-α-D-glucose 6-dehydrogenase inhibits glioblastoma growth and migration[J].Oncogene, 2018, 37(20):2615-2629.doi: 10.1038/s41388-018-0138-y.
[5] Wang TP, Pan YR, Fu CY, et al.Down-regulation of UDP-glucose dehydrogenase affects glycosaminoglycans synthesis and motility in HCT-8 colorectal carcinoma cells[J].Exp Cell Res, 2010,316(17):2893-2902.doi: 10.1016/j.yexcr.2010.07.017.
[6] Huang D, Casale GP, Tian J, et al.Udp-glucose dehydrogenase as a novel field-specific candidate biomarker of prostate cancer[J].Int J Cancer, 2010, 126(2):315-327.doi: 10.1002/ijc.24820.
[7] Arnold JM, Gu F, Ambati CR, et al.UDP-glucose 6-dehydrogenase regulates hyaluronic acid production and promotes breast cancer progression[J].Oncogene, 2019.doi: 10.1038/s41388-019-0885-4.[Epub ahead of print]
[8] Theocharis AD, Tsara ME, Papageorgacopoulou N, et al.Pancreatic carcinoma is characterized by elevated content of hyaluronan and chondroitin sulfate with altered disaccharide composition[J].Biochim Biophys Acta, 2000, 1502(2):201-206.doi: 10.1016/s0925-4439(00)00051-x.
[9] Toole BP.Hyaluronan: from extracellular glue to pericellular cue[J].Nat Rev Cancer, 2004, 4(7):528-539.doi: 10.1038/nrc1391.
[10] Hagiuda D, Nagashio R, Ichinoe M, et al.Clinicopathological and prognostic significance of nuclear UGDH localization in lung adenocarcinoma[J].Biomed Res, 2019, 40(1):17-27.doi: 10.2220/biomedres.40.17.
[11] Rhodes DR, Yu J, Shanker K, et al.ONCOMINE: a cancer microarray database and integrated data-mining platform[J].Neoplasia, 2004, 6(1):1-6.doi: 10.1016/s1476-5586(04)80047-2.
[12] Chandrashekar DS, Bashel B, Balasubramanya SAH, et al.UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses[J].Neoplasia, 2017, 19(8):649-658.doi: 10.1016/j.neo.2017.05.002.
[13] Kassambara A, Kosinski M, Biecek P, et al.survminer: Drawing Survival Curves using 'ggplot2'[EB/OL].https://rdrr.io/cran/survminer/.
[14] Yu G, Wang L G, Han Y, et al.clusterProfiler: an R package for comparing biological themes among gene clusters[J].OMICS,2012, 16(5):284-287.doi: 10.1089/omi.2011.0118.
[15] Mostafavi S, Ray D, Warde-Farley D, et al.GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function[J].Genome Biol, 2008, 9(Suppl 1):S4.doi:10.1186/gb-2008-9-s1-s4.
[16] Zhou Y, Zhou B, Pache L, et al.Metascape provides a biologistoriented resource for the analysis of systems-level datasets[J].Nat Commun, 2019, 10(1):1523.doi: 10.1038/s41467-019-09234-6.
[17] Huang WY, Yue L, Qiu WS, et al.Prognostic value of CRM1 in pancreas cancer[J].Clin Invest Med, 2009, 32(6):E315.
[18] Ungefroren H, Kruse M L, Trauzold A, et al.FAP-1 in pancreatic cancer cells: functional and mechanistic studies on its inhibitory role in CD95-mediated apoptosis[J].J Cell Sci, 2001, 114(Pt 15):2735-2746.
[19] Razidlo GL, Burton KM, McNiven MA.Interleukin-6 promotes pancreatic cancer cell migration by rapidly activating the small GTPase CDC42[J].J Biol Chem, 2018, 293(28):11143-11153.doi:10.1074/jbc.RA118.003276.
[20] DeBerardinis RJ, Lum JJ, Hatzivassiliou G, et al.The biology of cancer: metabolic reprogramming fuels cell growth and proliferation[J].Cell Metab, 2008, 7(1):11-20.doi: 10.1016/j.cmet.2007.10.002.
[21] Liu C, Karam R, Zhou Y, et al.The UPF1 RNA surveillance gene is commonly mutated in pancreatic adenosquamous carcinoma[J].Nat Med, 2014, 20(6):596-598.doi: 10.1038/nm.3548.
[22] Ide Y, Miyoshi E, Nakagawa T, et al.Aberrant expression of N-acetylglucosaminyltransferase-IVa and IVb (GnT-IVa and b)in pancreatic cancer[J].Biochem Biophys Res Commun, 2006,341(2):478-482.doi: 10.1016/j.bbrc.2005.12.208.
[23] Daemen A, Peterson D, Sahu N, et al.Metabolite profiling stratifies pancreatic ductal adenocarcinomas into subtypes with distinct sensitivities to metabolic inhibitors[J].Proc Natl Acad Sci U S A,2015, 112(32):E4410-4417.doi: 10.1073/pnas.1501605112.
[24] Gründker C, Ernst J, Reutter MD, et al.Effective targeted chemotherapy using AEZS-108 (AN-152) for LHRH receptorpositive pancreatic cancers[J].Oncol Rep, 2011, 26(3):629-635.doi: 10.3892/or.2011.1340.
[25] Griffin N, Faulkner S, Jobling P, et al.Targeting neurotrophin signaling in cancer: The renaissance[J].Pharmacol Res, 2018,135:12-17.doi: 10.1016/j.phrs.2018.07.019.
[26] Sheng W, Chen C, Dong M, et al.Calreticulin promotes EGFinduced EMT in pancreatic cancer cells via Integrin/EGFR-ERK/MAPK signaling pathway[J].Cell Death Dis, 2017, 8(10):e3147.doi: 10.1038/cddis.2017.547.
[27] Shen T, Cai L D, Liu YH, et al.Ube2v1-mediated ubiquitination and degradation of Sirt1 promotes metastasis of colorectal cancer by epigenetically suppressing autophagy[J].J Hematol Oncol, 2018,11(1):95.doi: 10.1186/s13045-018-0638-9.
[28] Tomasi ML, Ryoo M, Skay A, et al.Polyamine and methionine adenosyltransferase 2A crosstalk in human colon and liver cancer[J].Exp Cell Res, 2013, 319(12):1902-1911.doi: 10.1016/j.yexcr.2013.04.005.
[29] Yang H, Li TW, Peng J, et al.Insulin-like growth factor 1 activates methionine adenosyltransferase 2A transcription by multiple pathways in human colon cancer cells[J].Biochem J, 2011,436(2):507-516.doi: 10.1042/BJ20101754.
[30] Phuong NT, Kim SK, Im JH, et al.Induction of methionine adenosyltransferase 2A in tamoxifen-resistant breast cancer cells[J].Oncotarget, 2016, 7(12):13902-13916.doi: 10.18632/oncotarget.5298.
[31] Tian C, Jin Y, Shi S.Long non-coding RNA SUMO1P3 may promote cell proliferation, migration, and invasion of pancreatic cancer via EMT signaling pathway[J].Oncol Lett, 2018,16(5):6109-6115.doi: 10.3892/ol.2018.9378.
[32] Varshavsky A.The N-end rule pathway and regulation by proteolysis[J].Protein Sci, 2011, 20(8):1298-1345.doi: 10.1002/pro.666.
[33] Park JS, Lee JH, Lee YS, et al.Emerging role of LOXL2 in the promotion of pancreas cancer metastasis[J].Oncotarget, 2016,7(27):42539-42552.doi: 10.18632/oncotarget.9918.
[34] Paul D, Chanukuppa V, Reddy P J, et al.Global proteomic profiling identifies etoposide chemoresistance markers in non-small cell lung carcinoma[J].J Proteomics, 2016, 138:95-105.doi: 10.1016/j.jprot.2016.02.008.
[35] Yuan TL, Cantley LC.PI3K pathway alterations in cancer:variations on a theme[J].Oncogene, 2008, 27(41):5497-5510.doi:10.1038/onc.2008.245.
